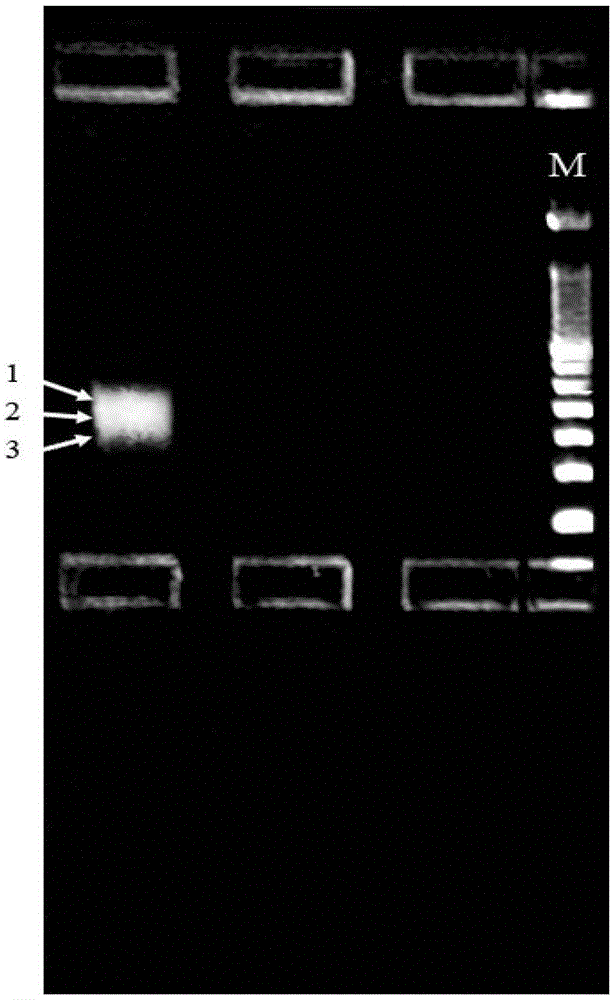
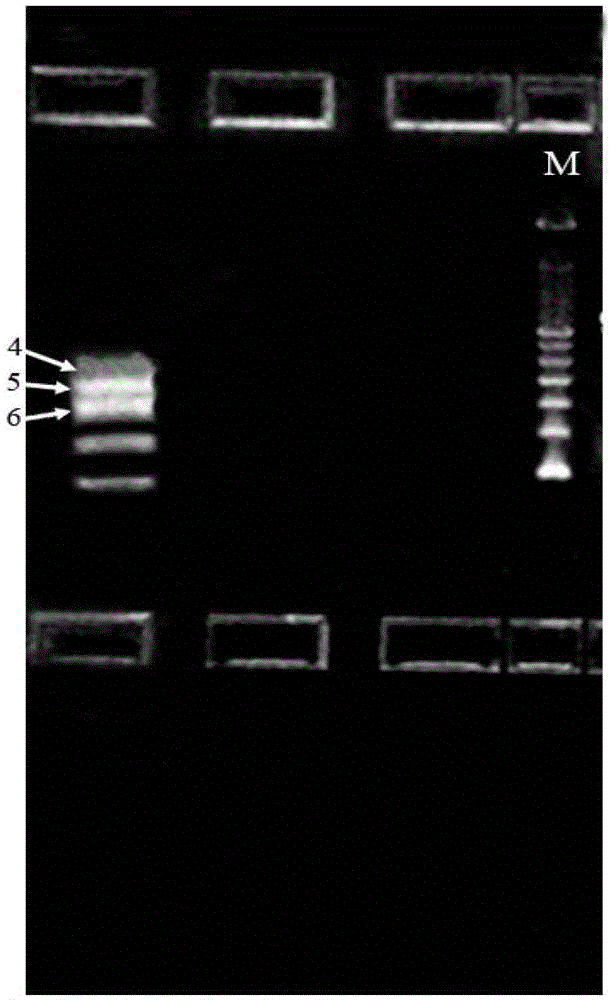
Asymmetric high-throughput sequencing linkers capable of effectively improving library construction efficiency, and application of linkers
A sequencing adapter, high-throughput technology, applied in the field of high-throughput sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] 1. Example 1: Construction of Tongue Cancer Genome 150bp Library
[0053] 1. Genomic DNA fragmentation (using CovarisS220 ultrasonic breaker)
[0054] (1) Take 2 μg of tongue cancer genomic DNA, and perform ultrasound interruption according to the set parameters, PeakincidentPower: 175, Dutyfactor: 10%, CyclesperBurst: 200, TreatmentTime(s): 430, Temperature(°C): 7, Waterlevel: 12, Sample Volume (μL): 130.
[0055] 2. 2% agarose gel electrophoresis (fragment selection)
[0056] (1) According to the DL500 molecular beacon, select fragments with a fragment size of 140-160bp for gel cutting and recovery. The agarose gel recovery kit used here is Tiangen agarose gel recovery kit;
[0057] 3. DNA fragments are subjected to end repair (the end repair kit used here is EpicentreEnd-it TM DNAEnd-RepairKit)
[0058] (1) Take 50ng of fragmented DNA of the above size for end repair: Fragmented DNA: 34 μL, 10×End-it buffer: 5 μL, End-itATP (10mM): 5 μL, End-itdNTPs (2.5mM): 5 μ...
Embodiment 2
[0081] 2. Example 2: Construction of Human Blood Circulating Nucleic Acid Library
[0082] 1. DNA fragment end repair
[0083] (1) Take 100 ng of circulating nucleic acid for end repair (the end repair kit used here is EpicentreEnd-itTMDNAEnd-RepairKit): Fragmented DNA: 34 μL, 10×End-it buffer: 5 μL, End-itATP (10 mM): 5 μL, End-itdNTPs (2.5mM): 5μL, End-itenzymemix: 1μL, Nuclease-freeWater: 0μL;
[0084] (2) Incubate at room temperature for 30 minutes;
[0085] (3) Purify the obtained product with magnetic beads, and the magnetic beads used here are BeckmanAgencourtAMPureXP nucleic acid purification magnetic beads.
[0086] 2. Add A to the 3' end of the DNA fragment that has been repaired
[0087] (1) Add A to the product (32 μL) obtained in the previous step: DNA: 32 μL, NEBBuffer: 25 μL, dATP (10 mM): 1 μL, Nuclease-freeWater: 9 μL, Klenowexo - : 3 μL;
[0088] (2) Incubate at 37°C for 30 minutes;
[0089] (3) Purify the obtained product with magnetic beads, and the m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com