Method for constructing high-flux and low-cost Fosmid library, label and label joint used in method
A tag and library technology, applied in the field of nucleic acid sequencing, high-throughput and low-cost Fosmid library construction, can solve the problems of laborious and time-consuming purification steps and loss of DNA, so as to reduce sequencing costs, reduce purification steps, and save library construction. effect of time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
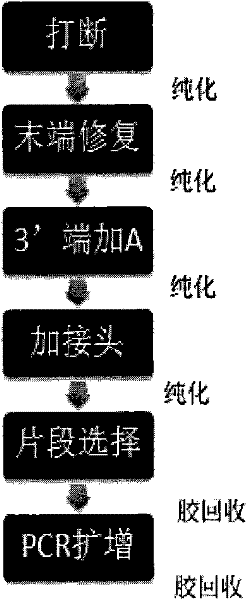
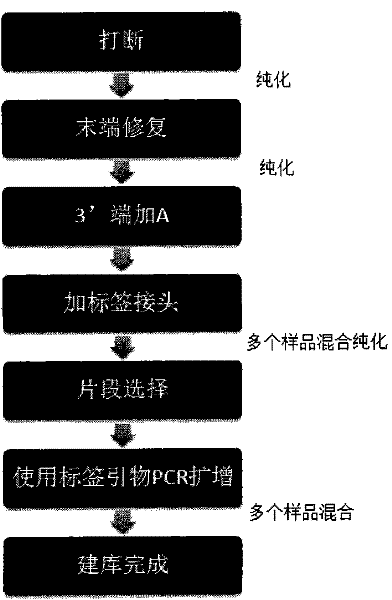
[0059] 1. Use four 96-well plates, respectively named plate No. 1, plate No. 2, plate No. 3, and plate No. 4. Each well contains 40 ul of 50 ng / ul Fosmid sample, and use the E210 model breaker produced by Covaris to break , the interrupt parameters are as follows:
[0060]
[0061] 2. Purify after interruption, take out 40ul from each well of the four 96-well plates into a new 96-well plate, recover and purify with Backman’s Ampure magnetic beads, and finally dissolve the samples in each well in 45ul ultrapure water, and take 42ul carry out the subsequent reaction.
[0062] 3. For end repair, configure the enzyme digestion reaction system in each well of four 96-well plates according to the following table:
[0063]
[0064] Place on a PCR instrument at 20°C for 30 minutes, Ampure magnetic beads are purified and dissolved in 22ul ultrapure water, and 17.7ul is taken out from each well and placed in a new 96-well plate for the following reaction.
[0065] 4. Add "A" bas...
Embodiment 2
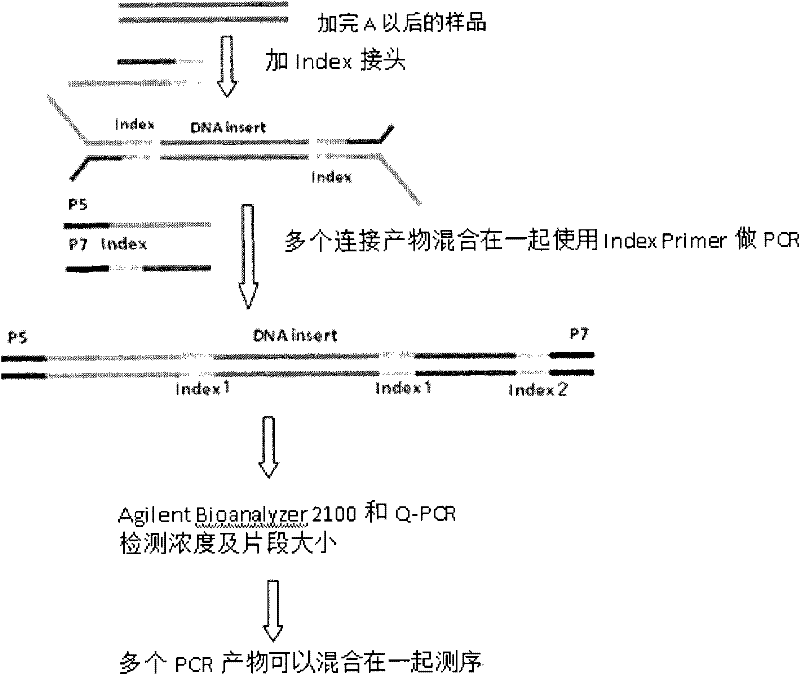
[0090] Illumina HiSeq Sequencing
[0091] According to the Q-PCR detection results, eight PCR products with a fragment size of 800 bp were combined in equal amounts and sequenced using the index PE101+8+101 cycle program. For details, see the Illumina HiSeq operating instructions. The results of the sequencing run are as follows:
[0092]
[0093] It can be seen from the results in the table that the deviation of the inserted fragment is less than 50bp, which meets the requirements, and the total data output is greater than 10G, indicating that the library was successfully built.
Embodiment 3
[0095] Result analysis
[0096] The sequencing results produced by Illumina HiSeq are a series of DNA sequences. By searching the adapter tag sequence, positive and negative primer tag sequences, and primer sequences in the sequencing results, the sequence information of the sample corresponding to each primer tag is established.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com