Method to Quantify siRNAs, miRNAs and Polymorphic miRNAs
a technology of mirnas and sirnas, applied in the field of methods to quantify sirnas, mirnas and polymorphic mirnas, can solve the problem of the silencing effect being a temporary phenomenon
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example
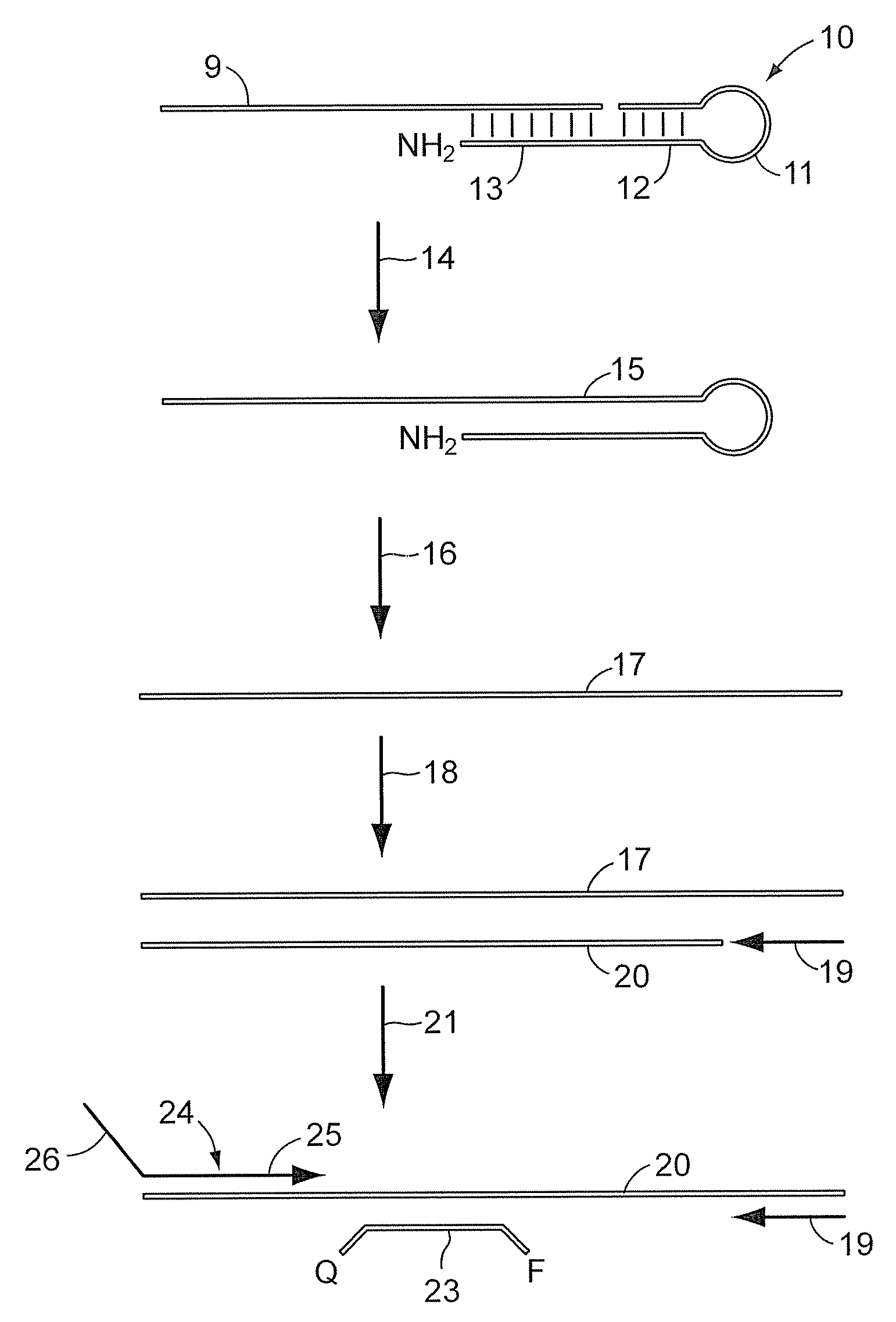
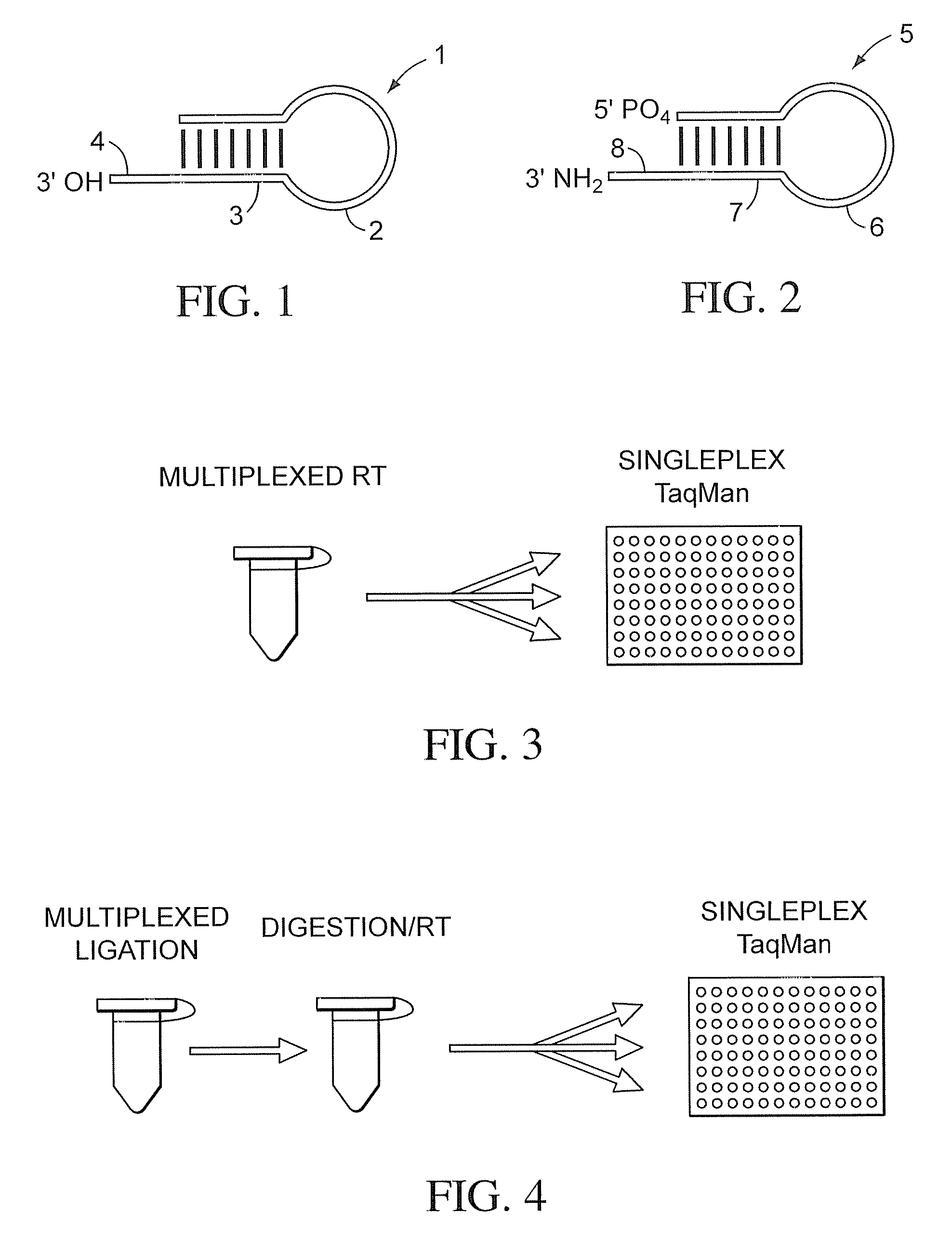
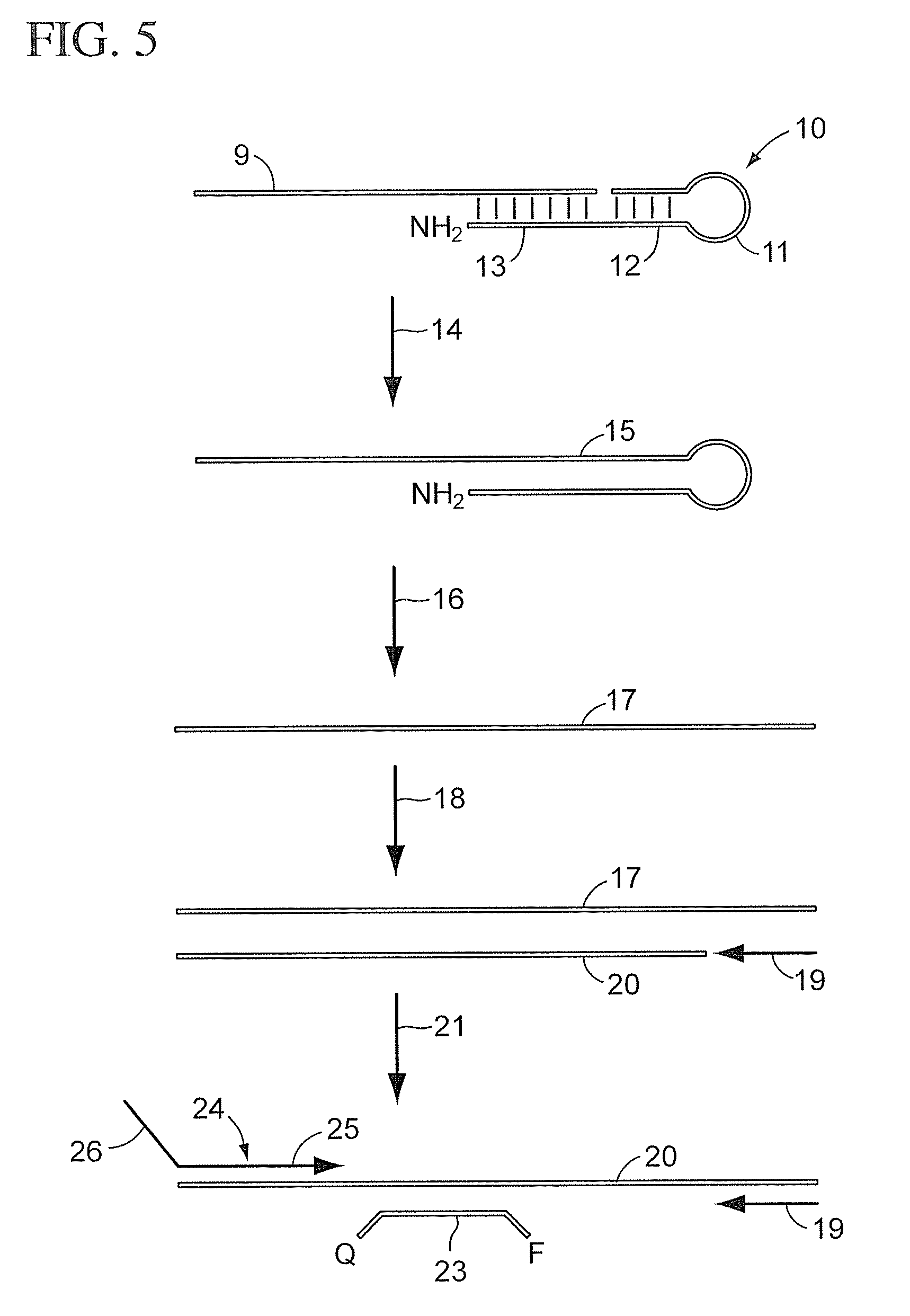
[0049]
TABLE 11. Ligation / kinaseVolumeStockReaction(uL)ItemsRNA sample 3×1×(12ng / ul)(4ng / ul)2.50reverse stem-loop ligation150nm50nm2.50probeLigation Reaction MixT4 polynucleotide kinase10U / uL0.5U / uL0.375(10 U / uL, NEB, M0201L)T4 DNA ligase (2000 U / uL,2000U / uL50U / uL0.188NEB, M0202M)T4 DNA Ligase buffer (NEB,10×1×0.750B02025)AB RNase Inhibitor (P / N:20U / uL0.5U / uL0.188N8080119)H2O1.000Total7.50
Incubate at 16° C. / 30 min, 37° C. / 60 min, 4° C. on hold.
[0050]
TABLE 22. DigestionstockvolumeItems(u / ul)MixtureReaction(ul)UNG (NEB, M0280L)20.40.10.1AB RNase Inhibitor200.50.125(P / N: N8080119)T4 DNA Ligase buffer1010.25(NEB, B02025)H202.025Total2.50
Add 2.5 ul of UNG mix to 7.5 ul of ligation mix, 37° C. / 60 min, 4° C. on hold.
[0051]
TABLE 33. RTVolumeStock[1×](uL)ItemsRT primer5um1um3.00Ligation-digestion Product 3×1×5.00RT Enzyme MixdNTPs with dTTP (P / N:25mM0.25mM0.1508080261)eacheachMultiScribe Reverse50×3.33× 1.000Transcriptase (P / N: 4319983)10× RT Buffer (P / N: 4319981)10×1×1.500AB RNase Inhibit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| acid | aaaaa | aaaaa |
| polymorphic | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com