Primer-mediated cyclized constant-temperature nucleic acid rolling circle amplification method and kit
A rolling circle amplification and cyclization technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve the problem of not being able to be completed at one time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
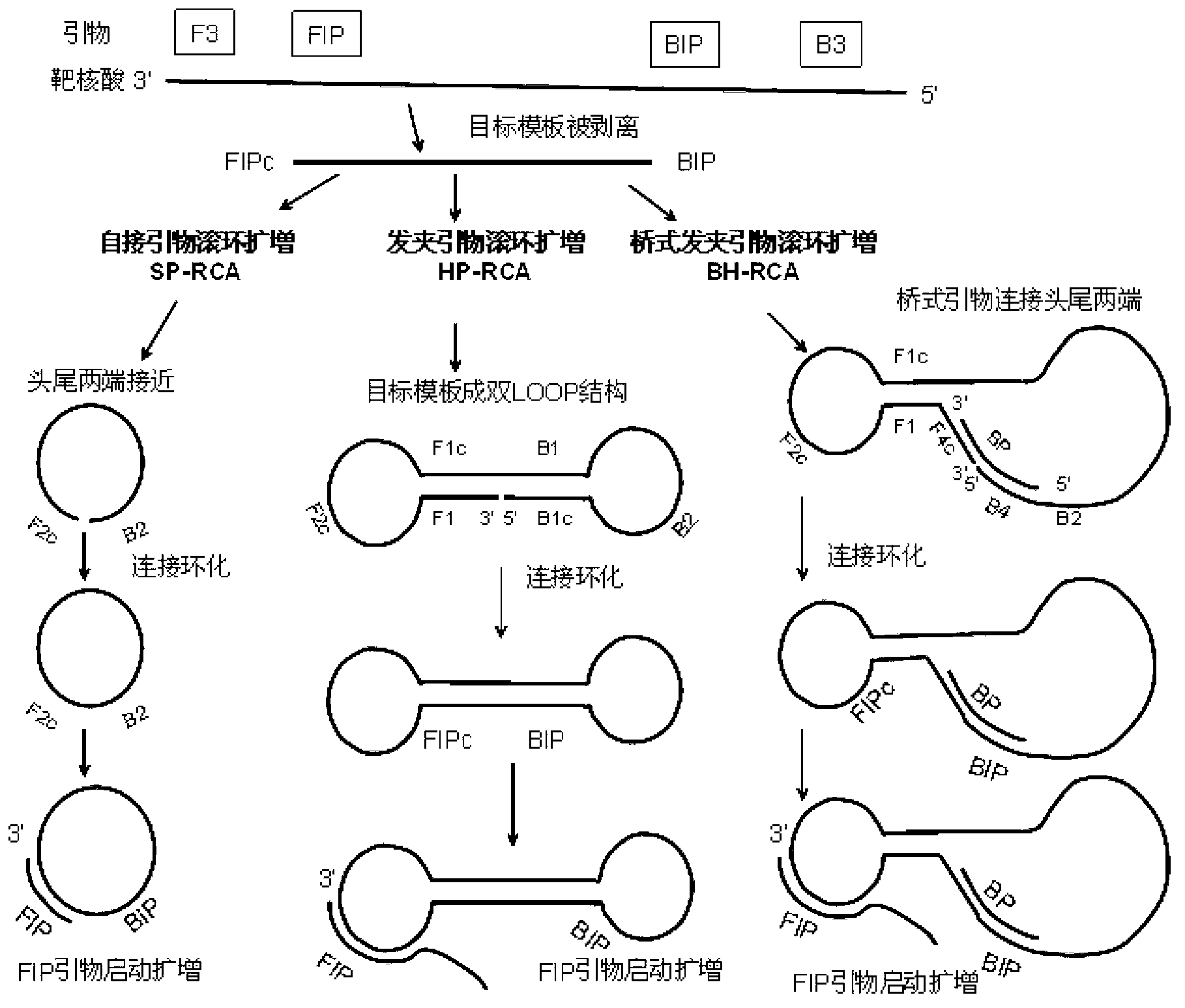
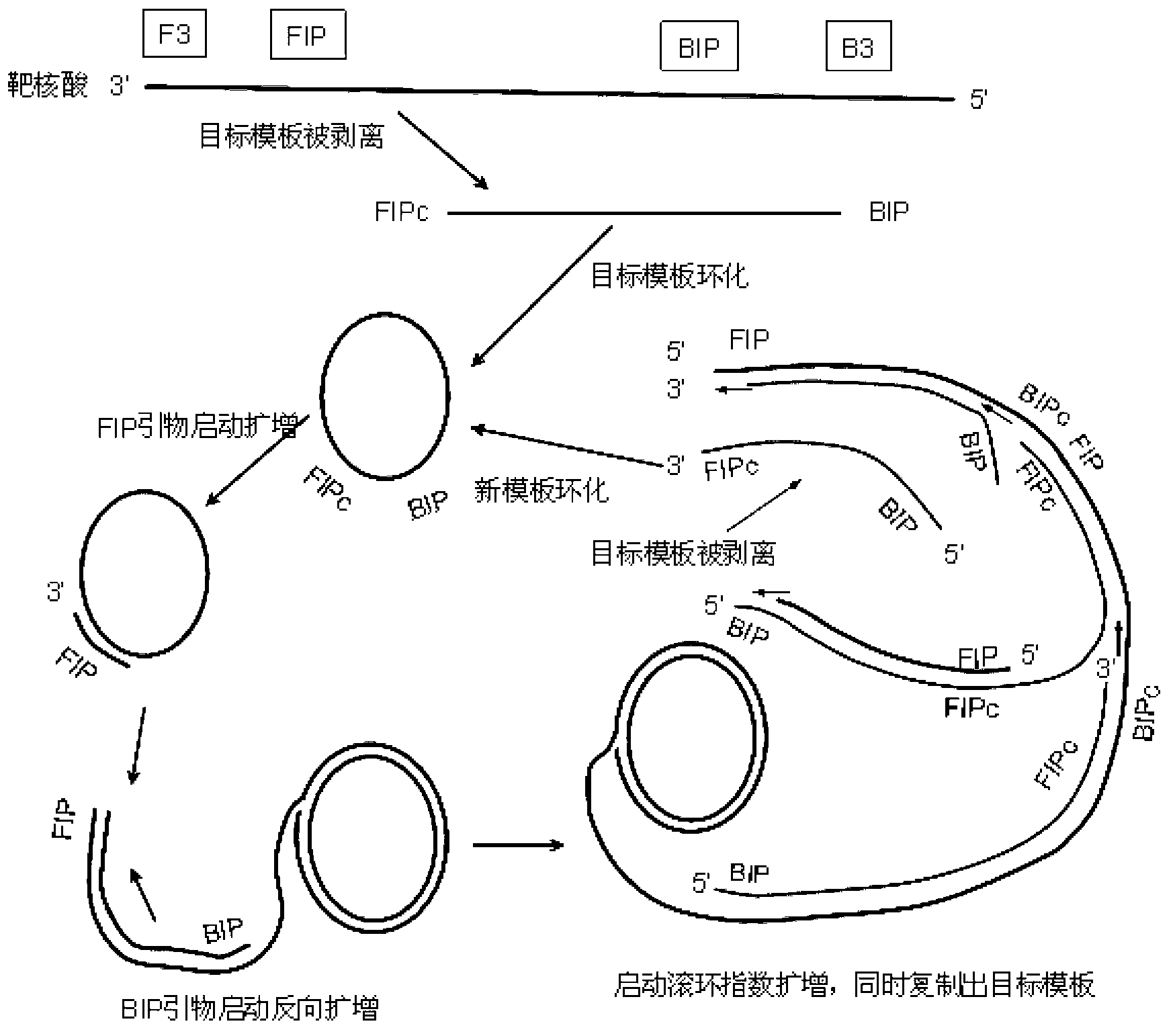
Image
Examples
Embodiment 1
[0078] Example 1: Bridge hairpin primer BHP-RCA detects double-stranded DNA
[0079] In this example, a bacterial culture of Listeria monocytogenes was used as a sample. Bacteria were purchased from ATCC (ATCC19116). Take 1 μL of the original seed solution into 3 ml of TSBYE medium, shake and culture at 35°C for 24 hours, take 200 μL of bacterial suspension, centrifuge at 5000 rpm for 2 minutes, and collect the bacteria for subsequent DNA extraction. According to the sequence information published by NCBI, the relatively conserved hemolysin coding gene hlyA of Listeria monocytogenes was used as the target sequence, and the partial sequence is as follows (GenBank NO.AB566375):
[0080] CAGATTTTTCGGCAAAGCTGTTACTAAAGAGCAGTTGCAAGCTTGGAGTGAATGCAGAAAATCCTCCTGCATATCTCCAAGTGT GGCATATGGCCGCCGTCCAAGTTTATTTGAAATTATCAACTAATTCCCATAGTACTAAAGTAAAAGCTGCTTTTGACGCTGCCGTAAGTGGGAAA TCTGTCTCAGGTGATGTAGAACTGACAAATATCAT (shown in SEQ ID NO. 1)
[0081] 1. According to the above sequences, design th...
Embodiment 2
[0109] Example 2: Bridge hairpin primer BHP-RCA detects single-stranded DNA
[0110] This implementation case uses porcine circovirus (Porcine Circovirus, PCV) inactivated vaccine as a sample. The vaccine was purchased from Harbin Veken Biotechnology Development Company, and the virus content of the vaccine was not less than 105.5 TCID50 before inactivation. PCV is a single-stranded closed circular DNA virus with a full-length genome of about 1.76 kb. PCV2 has high pathogenicity and is the main pathogen of post-weaning multisystemic wasting syndrome in piglets. According to the sequence information released by NCBI, the relatively conservative replication-related gene Rep of PCV2 is used as the target sequence, and the partial sequence is as follows (GenBank No. AF207700):
[0111] CTAGATCTCAAGGACAACGGAGTGACCTTGTCTACTGCTGTGAGTACCTTGTTGGAGAGCGGGAGTCTGGTGGCCGTTGCAGAGCAGC ACCCTGTAACGTTTGTCAGAAATTTCCGCGGGCTGGCTGAACTTTTGAAAGTGAGCGGGAAAATGCAGAAGCGTGATTGGAAGACGAATGT ACACGTCATTGTGGG...
Embodiment 3
[0135] Example 3: Bridge hairpin primer BHP-RCA detects RNA
[0136] In this embodiment, human immunodeficiency virus (Human Immunodeficiency Virus, HIV) is used as the detection object. Viral RNA samples were donated by Dr. Xu Feihai from Xiamen Wantai Canghai Biotechnology Co., Ltd. HIV is a single-stranded RNA retrovirus, and the vast majority of AIDS is caused by HIV-1. According to the sequence information published by NCBI, the gag gene, a relatively conserved structural protein of HIV-1, is used as the target sequence, and the partial sequence is as follows
[0137] (GenBank NO.: AF128998):
[0138] TCGACGGAGGACTCGGCTTGCTGAAGCGCGCACGGCAAGAGGCGAGGGGCGGCGACTGGTGAGTACGCCAAAATTTTTACTAGCGGAGGC TAGAAGGAGAGAGATGGGTGCGAGAGCGTCAGTATTAAGCGGGGGAAAATTAGATAAATGGGAGAAAATTCGGTTAAGGCCAGGAGGAAAGQGAACATATCAGTAAAAACATAGTATGGGGTAGAGTAGTAAGNOAAACATAGTATGG
[0139] 1. According to the above sequence, design the required five primers, the primer sequence is as follows:
[0140] Outer prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com