Fluorescence detection kit and fluorescence detection method for deletion mutation of gene
A fluorescence detection and gene deletion technology, applied in the field of chemical and biological sensing, can solve the problems of high requirements on experimental conditions, limited wide application, unsuitable for on-site use, etc., and achieves the effects of high sensitivity, simple operation and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
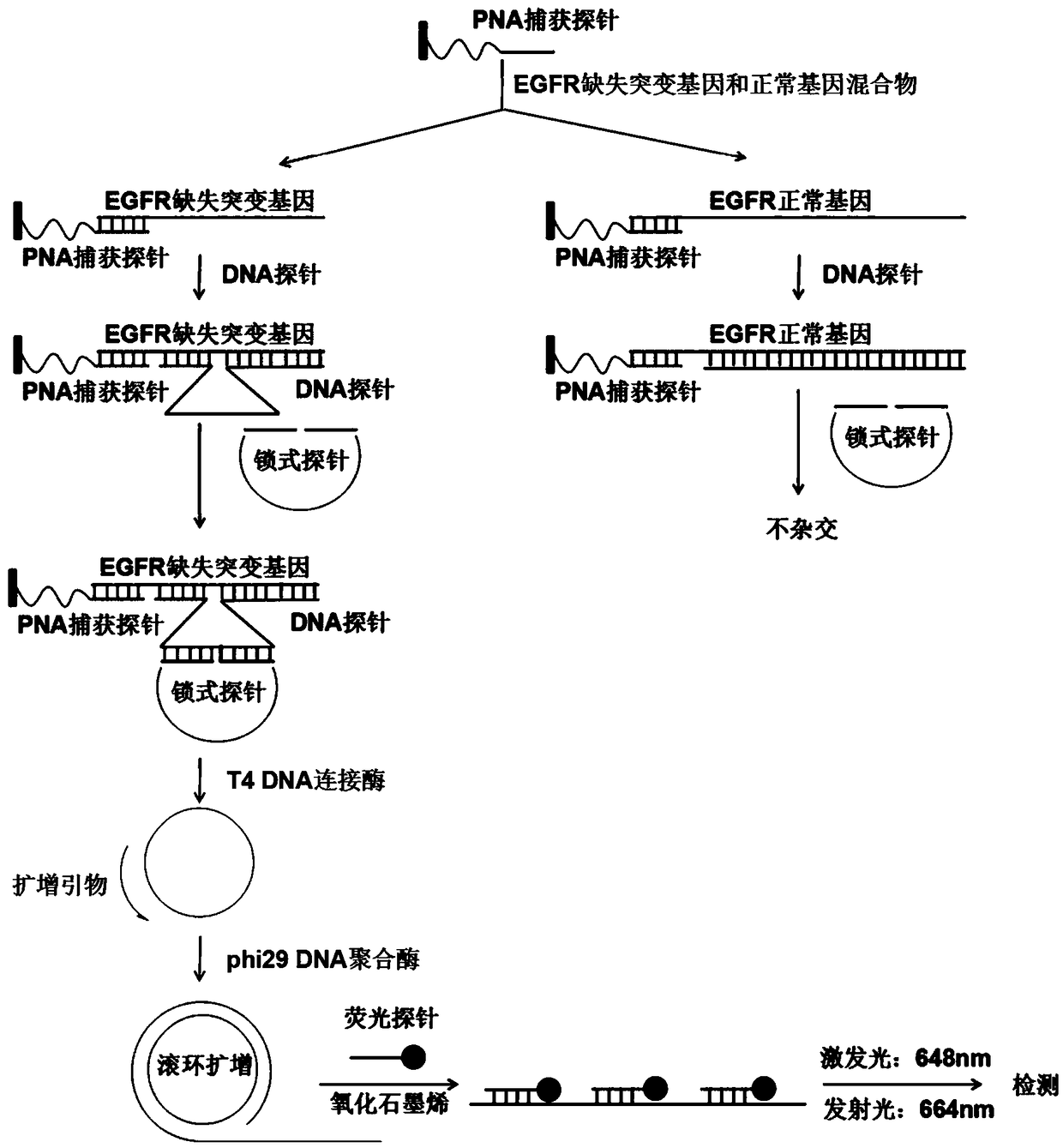
[0053] Design and synthesis of PNA capture probes, DNA probes, padlock probes, fluorescent probes and primers for rolling circle amplification
[0054] Select the 10-25 nucleotide sequence near the 3' end near the EGFR gene deletion site as the object, and design a PNA sequence that is completely complementary to it as a PNA capture probe, and the carboxyl terminal (CONH 2 ) is complementary to the 5' terminal base of the EGFR deletion mutant gene, and the amino terminal (NH 2 ) complementary base pairing with the 3' end of the EGFR deletion mutant gene, introducing 1-5 mPEGs at the carboxyl end to increase water solubility, using MBHA resin to condense one by one according to the base sequence under the conditions of HBTU and DIEA, after cleavage, purification and structural characterization For subsequent detection; DNA probes, padlock probes and rolling circle amplification primers were synthesized by Shanghai Jieli Biotechnology Co., Ltd. The detailed base sequences are sh...
Embodiment 2
[0058] Fluorescence detection of EGFR deletion mutation gene
[0059] (1) Immobilization of PNA capture probes on 96-well plate
[0060] Add 1 μL of 100 μM PNA capture probe and 100 μL NaHCO to each well 3 Fixative solution, the final concentration of PNA capture probe is 1.0μM, incubate with shaking (600-700rpm) at room temperature for 2 hours, then add 50μL of lysine shielding solution, continue shaking at room temperature (600-700rpm) for 0.5 hours, use 300μL 1× Wash four times with PBST, wash four times with 250 μL 1×PBS, store 250 μL 1×PBS for later use, remove 250 μL 1×PBS when in use, add 200 μL BLBs blocking solution, shake at room temperature (600-700 rpm) for 0.5 hours, and then remove the blocking solution , directly used in the next step of detection;
[0061] (2) Capture of EGFR deletion mutant gene
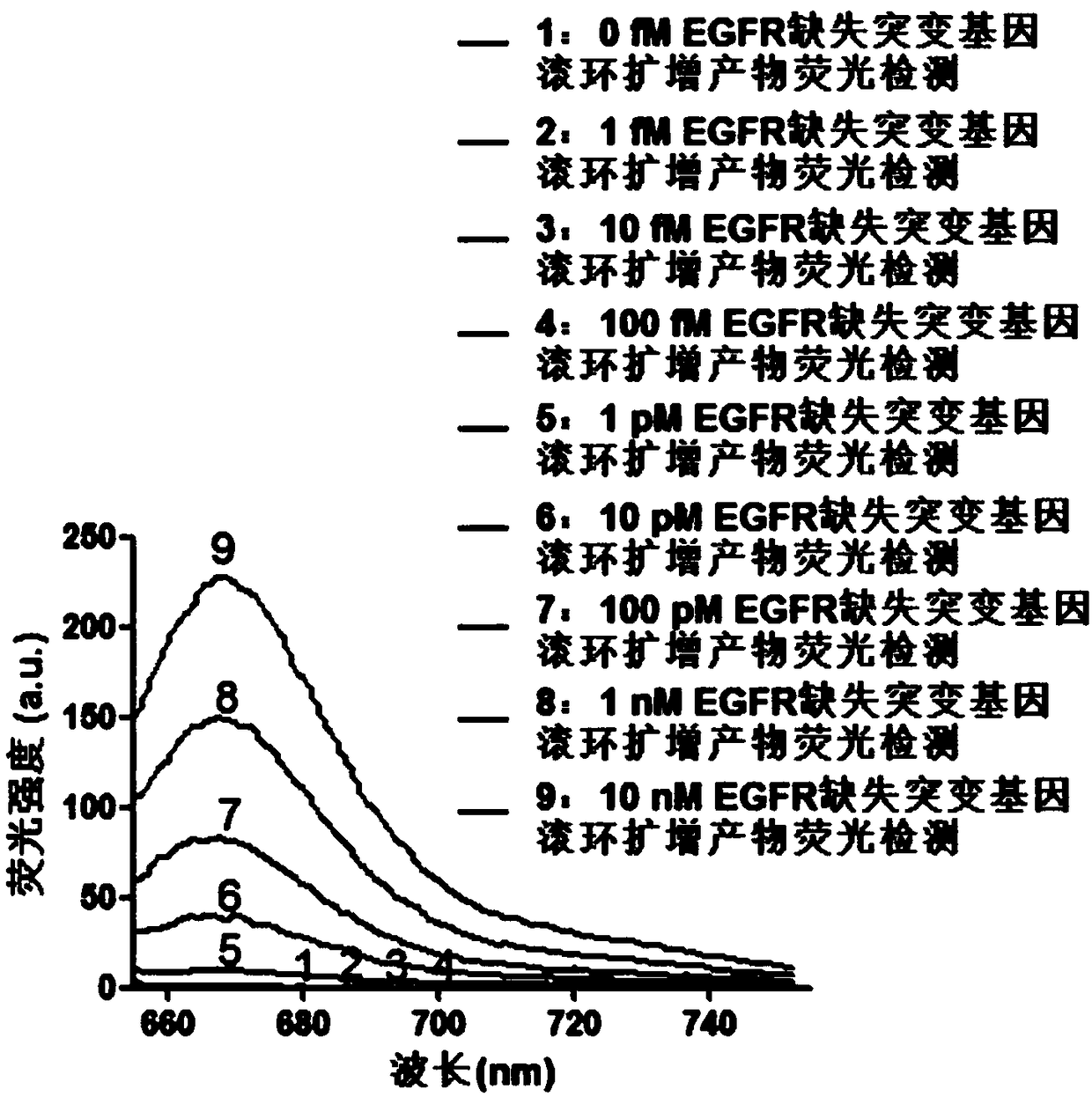
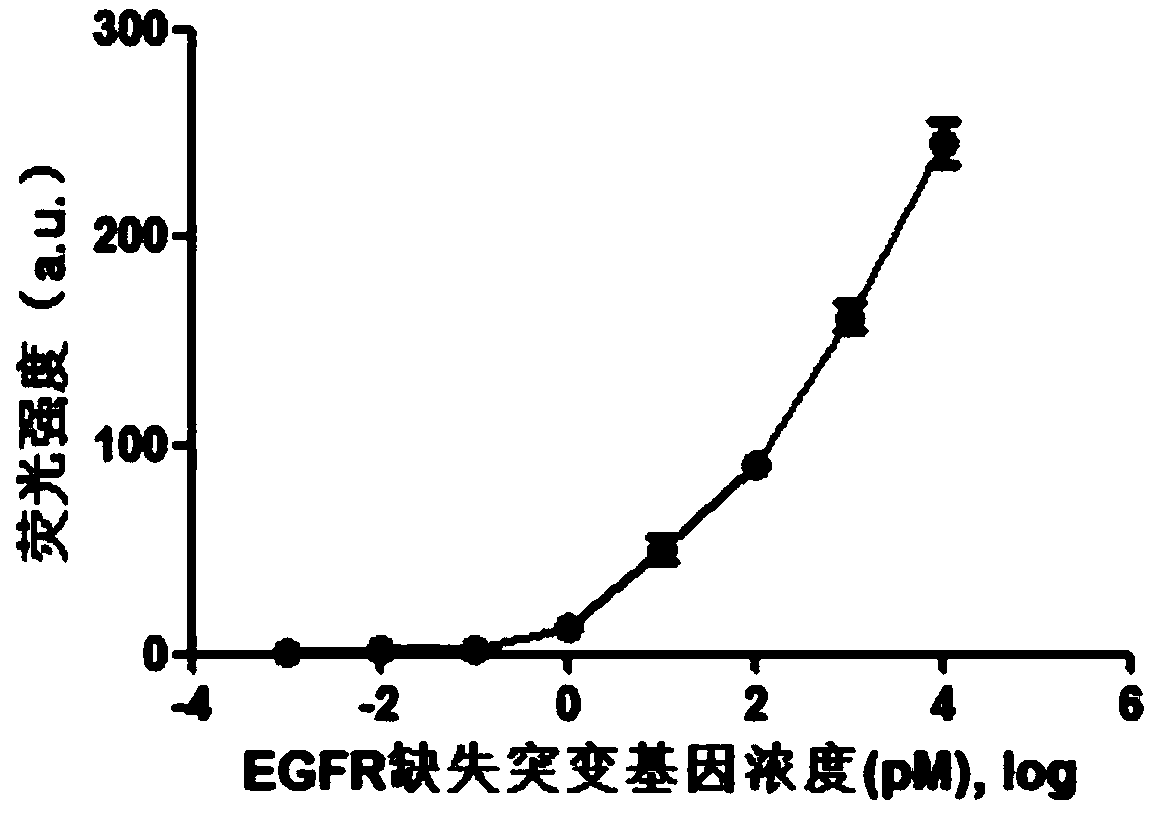
[0062] Add EGFR deletion mutant gene and 1×PBS buffer to 100 μL system to configure 9 concentration gradients (0fM, 1fM, 10fM, 100fM, 1pM, 10pM, 100pM, 1nM, 10nM)...
Embodiment 3
[0074] Negative control for fluorescence detection of EGFR deletion mutation gene
[0075] (1) Immobilization of PNA capture probes on 96-well plate
[0076] Add 1 μL of 100 μM PNA capture probe and 99 μL NaHCO to each well 3 Fixative solution, the final concentration of PNA capture probe is 1.0μM, incubate with shaking (600-700rpm) at room temperature for 2 hours, then add 50μL of lysine shielding solution, continue shaking at room temperature (600-700rpm) for 0.5 hours, use 300μL 1× Wash four times with PBST, wash four times with 250 μL 1×PBS, store 250 μL 1×PBS for later use, remove 250 μL 1×PBS when in use, add 200 μL BLBs blocking solution, shake at room temperature (600-700 rpm) for 0.5 hours, and then remove the blocking solution , directly used in the next step of detection;
[0077] (2) Capture of EGFR gene
[0078] The experiment was divided into 5 groups in a 96-well plate: a, b, c, d, e, wherein 1 μL of 1 μM EGFR deletion mutant gene was added to group a, c and ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com