Misgurnus anguillicaudatus and paramisgurnus dabryanus specie identification primer and identification method
A technology of paraloach and loach, applied in biochemical equipment and methods, microbe measurement/testing, DNA/RNA fragments, etc., to achieve rapid and efficient germplasm identification and solve species misjudgment effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1: The present invention provides a pair of primers based on differences in mitochondrial DNA sequences to identify two species of loach and paraloach. The primer sequences are:
[0019] Upstream primer: 5'-CCGCCCAGGAGTATTTTATG-3' (shown in SEQ ID NO.1);
[0020] Downstream primer: 5'-TGGCATTTCATCAGGGGTG-3' (shown in SEQ ID NO.2).
Embodiment 2
[0022] The present invention provides a method for identifying loach and paraloach based on the primers described in Example 1. In this example, the samples of loach were collected in Zhoushan, Zhejiang, and the samples of paraloach were collected in Qinhuangdao, Hebei. Species identification was carried out according to traditional morphological methods. DNA was extracted according to the conventional phenol / chloroform method, and the collected samples were placed in 400 μL of lysate (NaCl50mmol / L, Tris-Cl (pH=8.0) 30mmol / L, EDTA (pH8.0) 100mmol / L, 1% SDS, Proteinase K 200 μg / mL), incubated at 50°C until clarified, then extracted once with an equal volume of saturated phenol:chloroform:isoamyl alcohol (25:24:1), and then precipitated with an equal volume of isopropanol. Then wash the precipitate with 0.4mL 75% ethanol, dissolve it with TE, and store it at -20°C for future use. The samples were amplified by PCR using the primers described in Example 1. The PCR reaction syste...
Embodiment 3
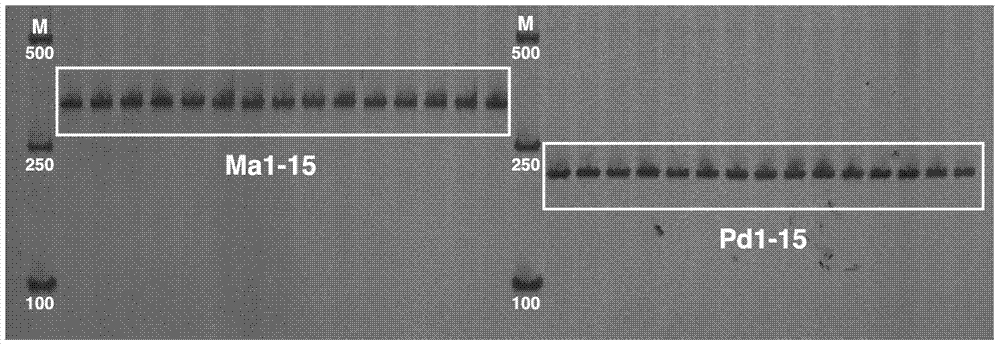
[0025] The method for identifying loach and paraloach as described in Example 2, the difference is that the samples of loach and paraloach in this embodiment were collected in Panjin, Liaoning, and the PCR amplification product was purified with 8% non-denaturing polymer Acrylamide gel electrophoresis detection, silver staining, scanner scanning record.
[0026] The results showed that a band of 285bp appeared in the 15 samples that were judged as loach by morphology; and a band of 214bp appeared in the 15 samples that were judged by morphology as Paraloach. The identification results of this method are consistent with those based on morphological judgment, which proves the reliability of this method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com