Method for identifying related species of oyster
A technology for species identification and oysters, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of distinguishing different species, difficult species identification, cumbersome process, etc., and achieves wide applicability, clear and intuitive results, and method simple and easy effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
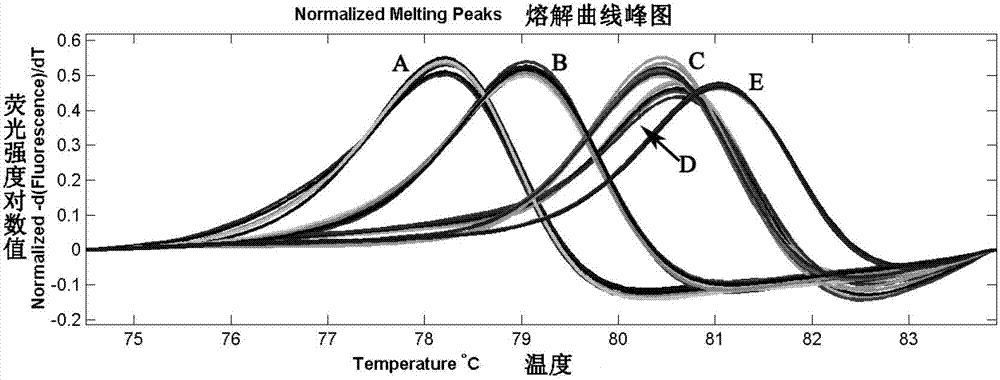
[0025] Identification of oyster relatives in the genus Pyrex:
[0026] There are many species of oysters in the genus Colossus, and the appearance of oysters is greatly affected by the environment, so it is difficult to correctly distinguish different species by phenotype. In terms of species classification and identification, there used to be many controversies. Nowadays, it is gradually tending to identify different species by using the difference of biomarker sequences such as mitochondrial COI gene sequence among different species.
[0027] a. Acquisition of the basis sequence for species identification: The COI gene sequences of different closely related oyster species in the genus Pyramid that are relatively common in my country are used as the basis sequence for species identification.
[0028] b. Primer design: Select a sequence region that can clearly distinguish the target species in the above-mentioned oyster COI gene, and use Primer Premier 5 software to design PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com