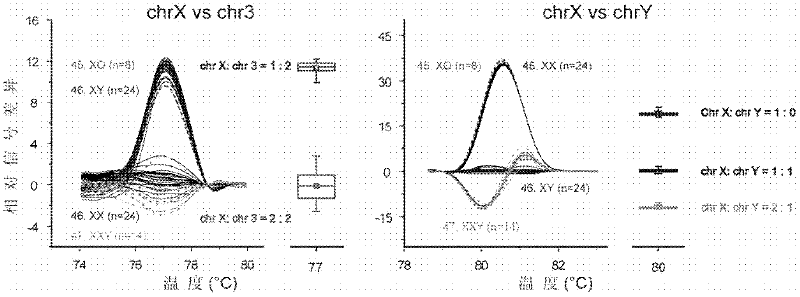
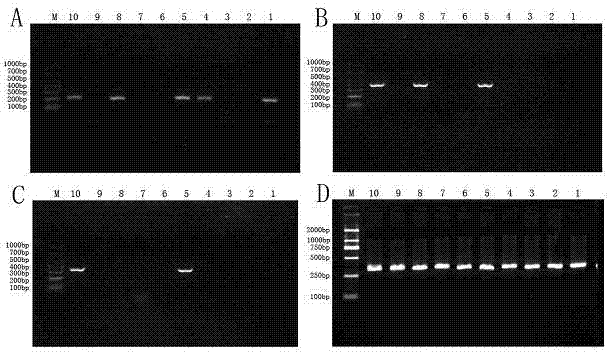
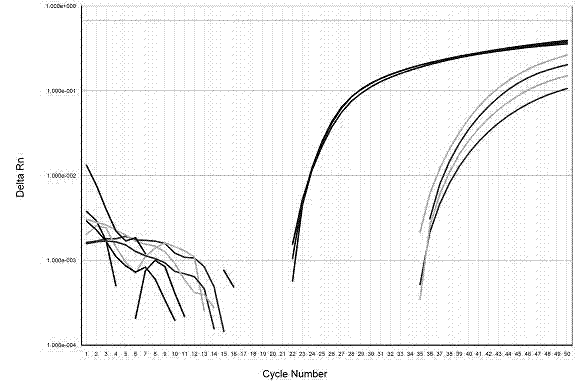
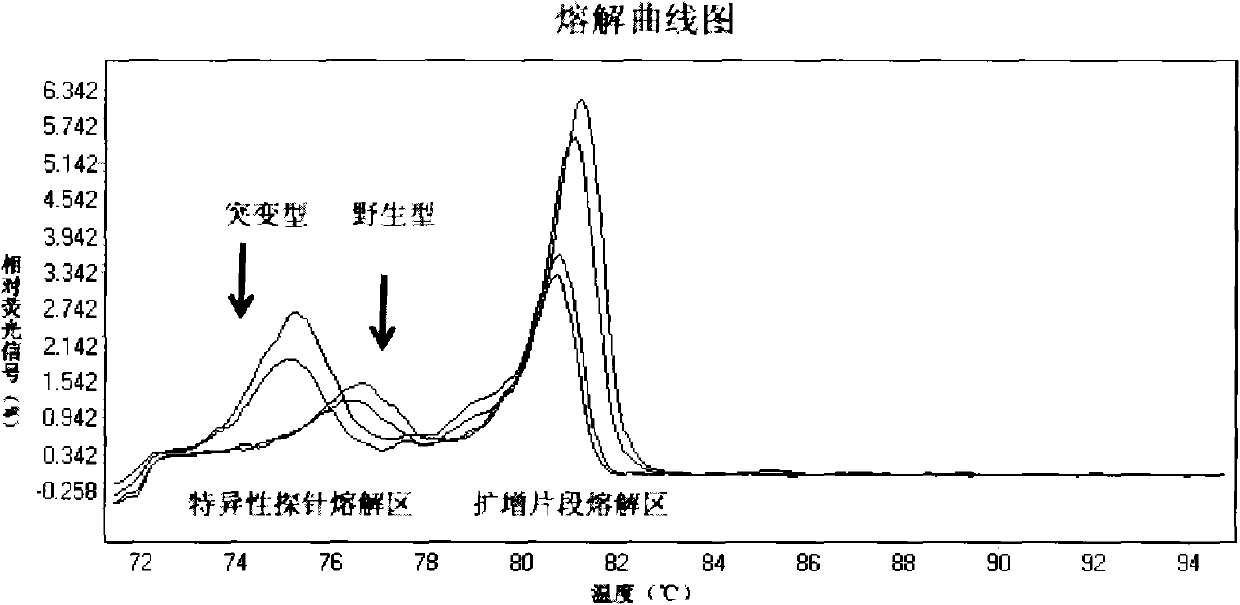
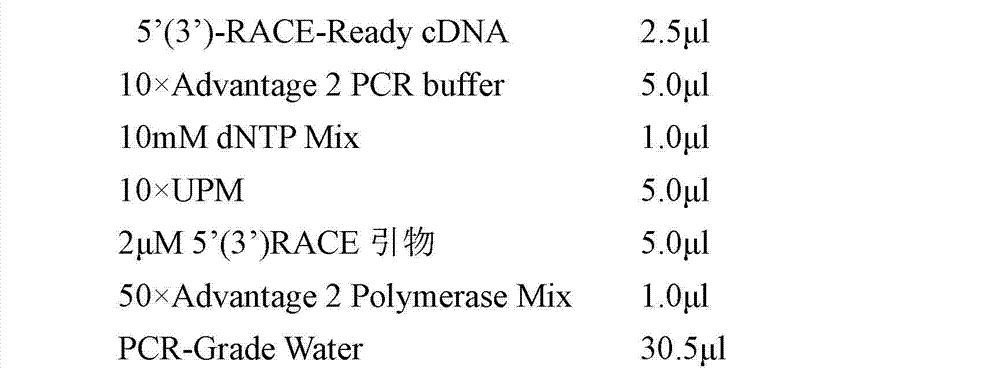
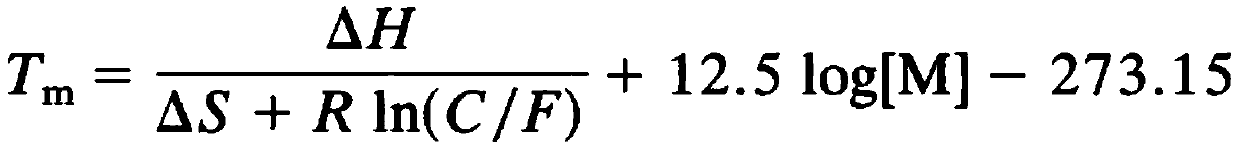Patents
Literature
152 results about "High Resolution Melt" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
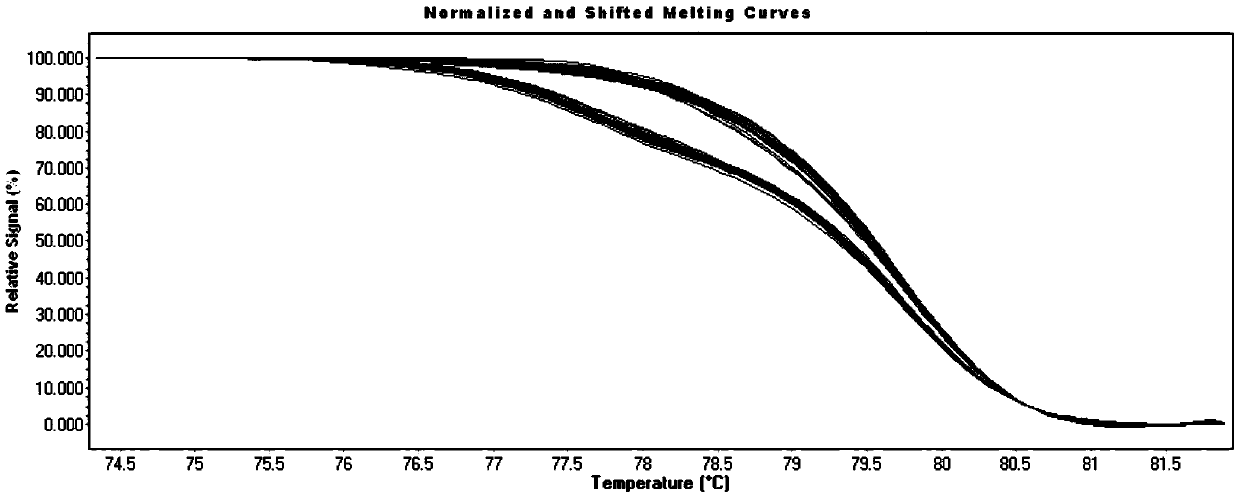
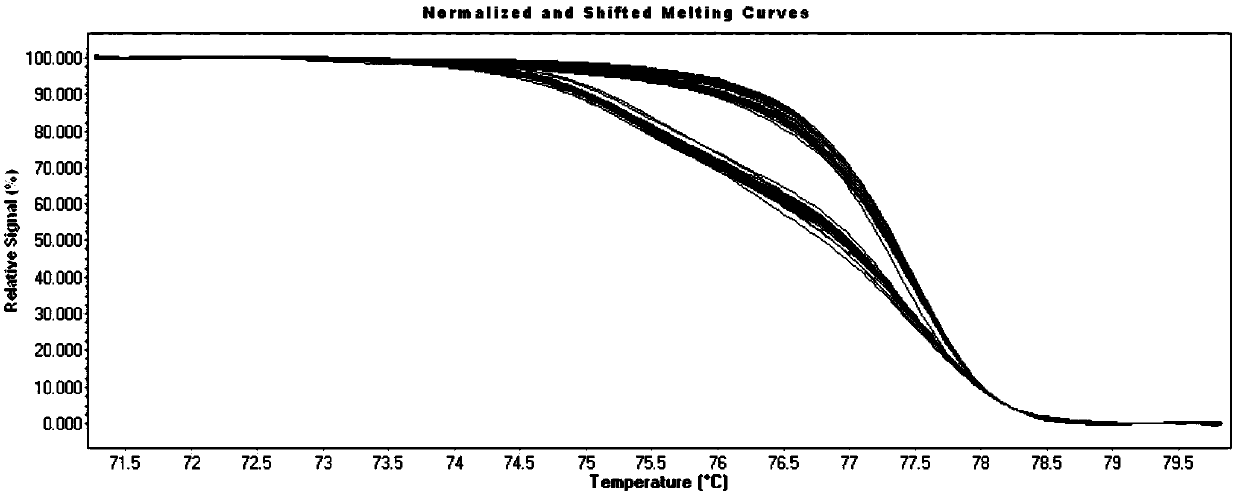
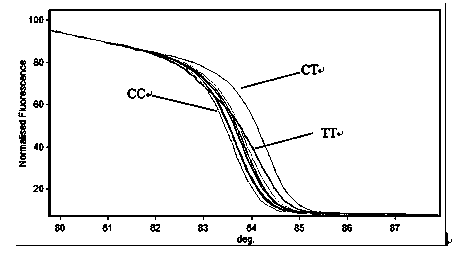
High Resolution Melt (HRM) analysis is a powerful technique in molecular biology for the detection of mutations, polymorphisms and epigenetic differences in double-stranded DNA samples. It was discovered and developed by Idaho Technology and the University of Utah.
Preparation method of non-transgenic CRISPR mutant
InactiveCN108611364AReduce frequencyImprove accuracyNucleic acid vectorVector-based foreign material introductionScreening methodSexual reproduction
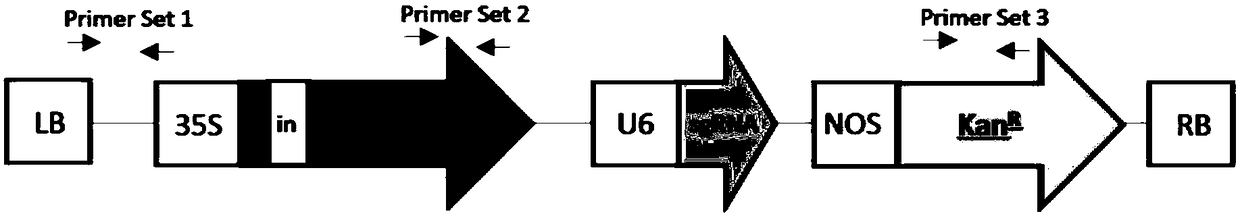
The invention provides a preparation method of a non-transgenic mutant based on a CRISPR-Cas9 technology. Specifically, the preparation method comprises a construction method and a screening method. The construction method is characterized in that CRISPR-Cas9 and sgRNA are preassembled and are located at T-DNA of agrobacterium tumefaciens; site-directed change of a target gene of a target plant can be realized by infection of the target plant by the agrobacterium tumefaciens and coculture without integrating the sequences of the CRISPR-Cas9 and the sgRNA into a genome of the target plant, so that an obtained site-directed mutant material of the target gene does not need the processes of sexual reproduction, segregation posterity, population screening and the like or does not contain any exogenous gene sequence. The obtained regeneration seedlings are screened by a high-throughput sequencing and high-resolution melting curve technology provided by the invention; mutant plants of which target genes are mutated can be obtained by identifying the regeneration seedlings efficiently and quickly at the current generation of transgene, even if low-proportion mutants of which the proportionis 1 / 100 of a mixed population can be screened. The screening method provided by the invention still can ensure excellent accuracy and sensitivity.
Owner:NANJING AGRICULTURAL UNIVERSITY
Method for detecting gene copy number variation
ActiveCN102409088ASimple designThe result is accurateMicrobiological testing/measurementPrenatal diagnosisWild type
Owner:郭奇伟 +1
Method used for detecting HLA-B*5801 alleles
ActiveCN103484533AReliable resultsFlexible detection methodMicrobiological testing/measurementDNA/RNA fragmentationGenomicsA-DNA
The invention belongs to the field of pharmacogenomics and genetic diagnosis, and relates to a method used for detecting HLA-B*5801 alleles. The method comprises following steps: a DNA sample to be detected is taken, three pairs of specific primers and a pair of internal primers are taken, amplification of DNA segments is realized by using sequence specific primer method, and then the results of the amplification are analyzed by agarose gel electrophoresis; or sample DNA is extracted, a pair of specific primers, a pair of internal primers and three fluorescence probes are taken, amplification of DNA segments is realized by Taqman probe method using a fluorescence ration PCR instrument, and then the amplification curve is analyzed so as to obtain results. Results analysis methods such as agarose gel electrophoresis, high resolution melting curve and SYBRGreen fluorogenic quantitative PCR are employed in the method. The method has advantages of speediness, convenience, flexibility, high resolution and no contamination; is suitable for detection of HLA-B*5801 alleles in samples such as peripheral blood and hair; and can be used for determining the probability of severe skin adverse reaction of patients with gout or hyperuricemia caused by taking of allopurinol.
Owner:安徽同科生物科技有限公司
Method and kit for detecting mutation of BRAF gene of human colorectal cancer
InactiveCN102586401ASignificant progressEasy to detectMicrobiological testing/measurementFluorescence/phosphorescenceNucleotideV600E
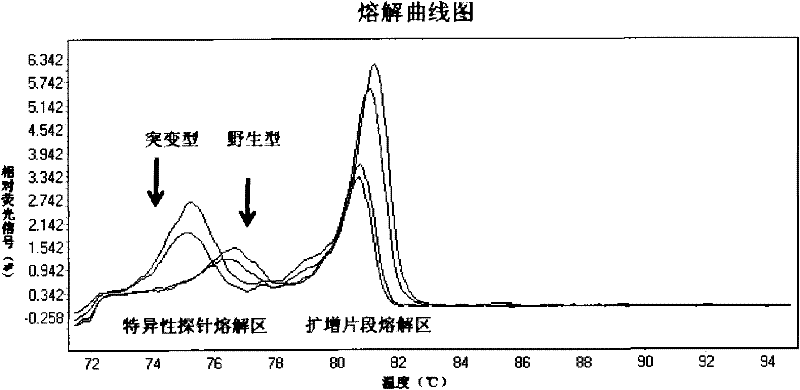
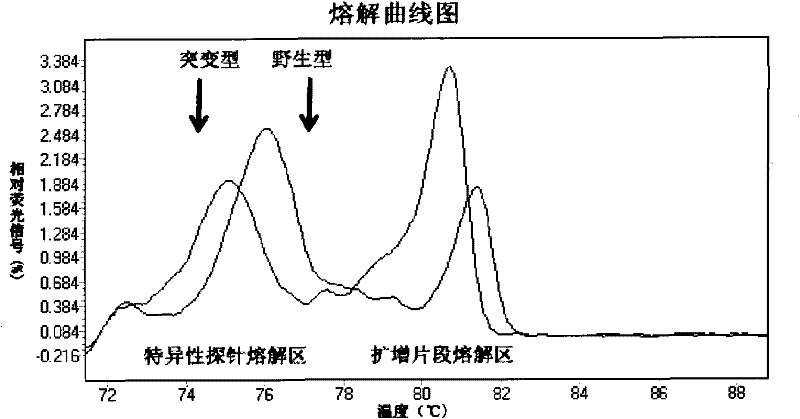
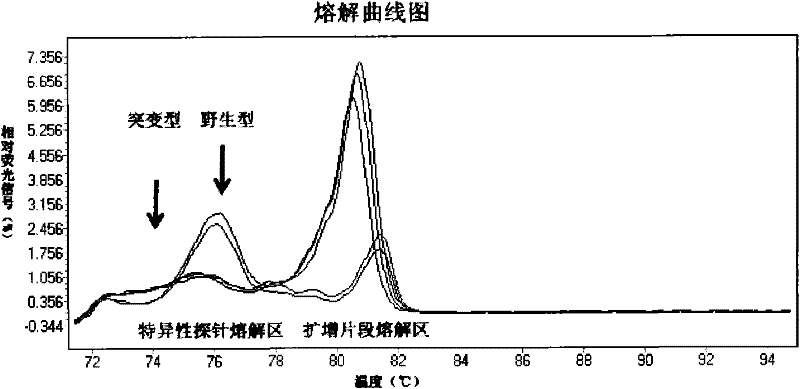
The invention relates to a method and a kit for detecting gene mutation, in particular to a method and a kit for detecting the mutation of BRAF gene. The invention is characterized in that the kit comprises a specific probe used for carrying out genotyping on No. 15 exon codon V600E of the BRAF gene, wherein the specific probe of the No. 15 exon codon V600E comprises a nucleotide sequence of V600E codon. by the technology combining the conventional polymerase chain reaction (PCR) amplification with a Cold-PCR enrichment amplification product and a high resolution melting curve analysis technology, the kit provided by the invention can be used for completing the judgment on sample genotyping.
Owner:苏州科贝生物技术有限公司
New system and kit for screening and diagnosing phenylketonuria
ActiveCN105755109ALow costImprove throughputMicrobiological testing/measurementGenes mutationImage resolution
The invention discloses a new system for screening and diagnosing phenylketonuria.41 common phenylalanine hydroxylase gene mutation sites and 4 most common 6-pyruvoyl tetrahydrobiopterin synthetase (PTPS) gene (PTS) mutation sites are screened to form the gene mutation detection system of phenylketonuria.The system is low in cost and high in throughput.Compared with the prior art, the system has the advantages that most mutation sites can be detected in one system, and 45 mutation sites can be detected; the comprehensive cost is the lowest, and the total cost of the system is lower than built methods using the DNA chip technology, the 'molecular switch' technology, the high-resolution melting curve method and the SNaPShot technology to detect PAH gene mutation; most mutation sites can be detected, the number of the detected mutation sites is twice as the number of the detected PAH gene mutation sites of the high-resolution melting curve method built by Xiamen University, the number of the detected mutation sites is 4 times as the number of the detected mutation sites of the DNA chip technology and the SNaPShot technology, and the number of the detected mutation sites is 6 times as the number of the detected mutation sites of the 'molecular switch' technology.
Owner:SUZHOU MUNICIPAL HOSPITAL +1
Method and kit for detecting human PIK3CA gene mutation
InactiveCN102533958ASignificant progressEasy to detectMicrobiological testing/measurementFluorescence/phosphorescenceMicrobiologyGenotyping
The invention relates to a method and kit for detecting gene mutation, and specifically relates to a method and kit for detecting PIK3CA gene mutation. The invention is characterized in that the kit comprises two specific probes for performing genotyping on a 9 exon and a 20 exon of the PIK3CA gene, wherein the specific probe for the 9 exon contains a nucleotide sequence of E545K and E542K codons, and the specific probe for the 20 exon contains a nucleotide sequence of an H1047R codon. By using the conventional PCR (polymerase chain reaction) amplification in combination with the Cold-PCR amplification product enrichment technology and the high-resolution melting curve analysis technology, the kit provided by the invention can complete the judgment on the genotyping of a sample.
Owner:苏州科贝生物技术有限公司
Method for identifying related species of oyster
ActiveCN102758009ASimple methodNo sequencing requiredMicrobiological testing/measurementOysterGenome
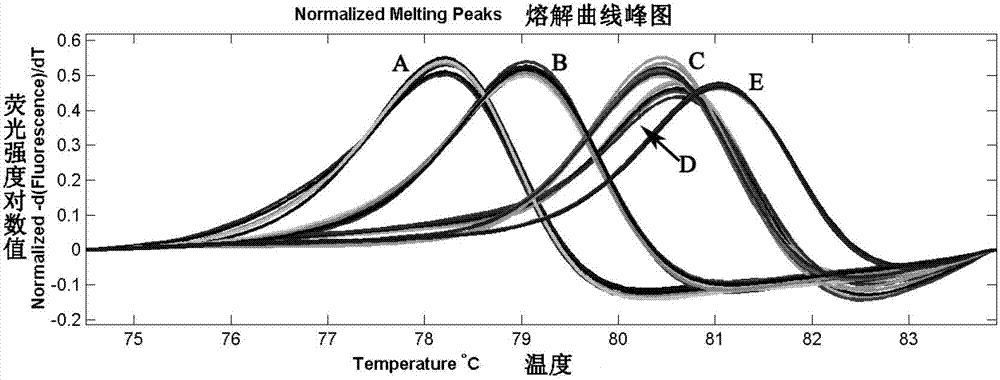
The invention relates to a related specie identification technology, in particular to a method for identifying related species of an oyster. The method specifically comprises the following steps of: by using an oyster genome DNA (Deoxyribonucleic Acid) as a template, designing a primer according to a tag sequence of the oyster specie genome; carrying out PCR (Polymerase Chain Reaction) amplification on the primer; and distinguishing oyster species by using Tm value difference in a high-resolution melting curve of an amplification product. Through the adoption of the method provided by the invention, the species can be identified rapidly, economically, simply and conveniently; and compared with a tag sequence sequencing result, an identifying result of the method provided by the invention is 100% in accuracy.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
SNP locus related to growth characteristics of patinopecten yessoensis and detection and application thereof
InactiveCN103740729AHigh trait valueAvoid crossbreedingMicrobiological testing/measurementClimate change adaptationInsulin-like growth factor bindingNucleotide sequencing
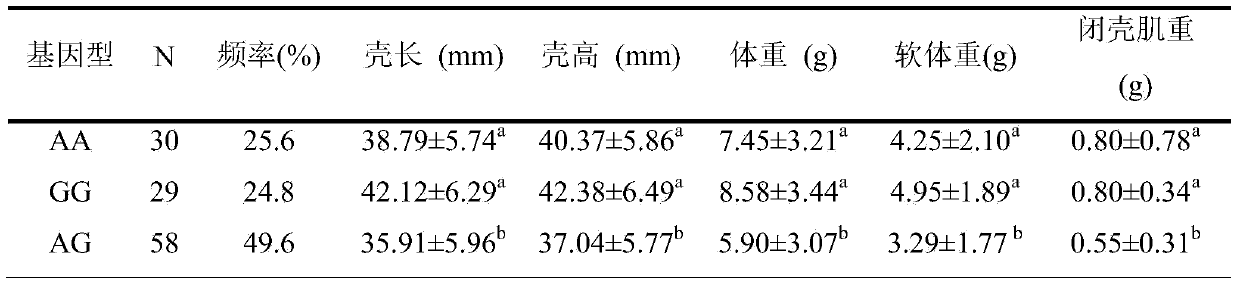
The invention provides an SNP locus related to growth characteristics of patinopecten yessoensis. The locus is the 1054 site of IGFBP (Insulin Like Growth Factor Binding Proteins) genes of which the nucleotide sequence is shown as SEQ ID NO:2 and has the base of A or G. The invention also provides a probe for detecting the SNP locus and a parting primer. The transcription sequence of the IGFBP genes in the patinopecten yessoensis is subjected to sequencing and Clustal comparison, and three SNP loci are screened. The site polymorphism is detected in the patinopecten yessoensis group by using a high resolution melting curve technology, and correlation between the site genotype frequency and the growth characteristics of patinopecten yessoensis is analyzed. The locus C.1054A>G is obviously related to important growth characteristics such as the height, shell length, weight, soft weight and adductor muscle weight of the patinopecten yessoensis, the growth characteristics of the AG type individuals are obviously lower than those of AA and GG type individuals (P is less than 0.05), and the characteristic value of the GG type is the highest. Therefore, individuals of which the genotype is GG on the locus can be preferentially selected during production and are used as parents for performing high-yield patinopecten yessoensis breeding or performing large-scale breeding.
Owner:OCEAN UNIV OF CHINA
Method for screening rice plant subjected to targeted gene editing
ActiveCN105154566AEliminate electrophoresisSimple and fast operationMicrobiological testing/measurementGenome editingFluorescence
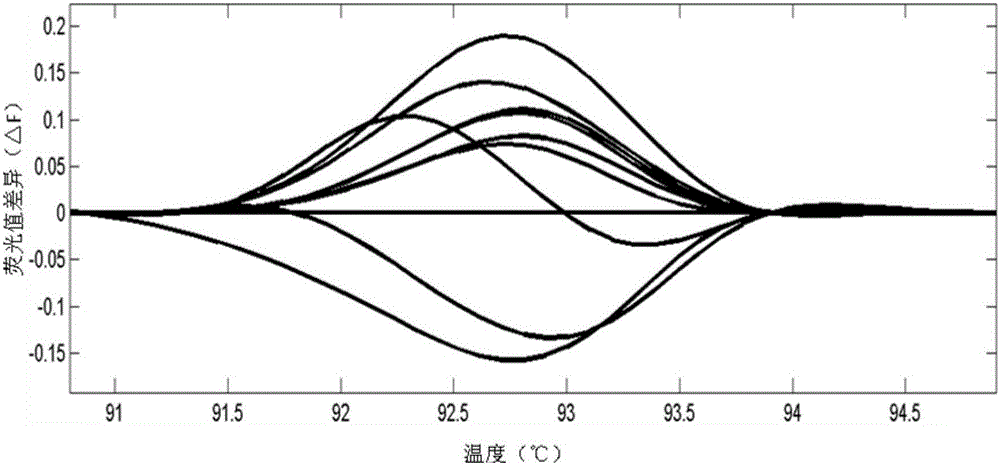
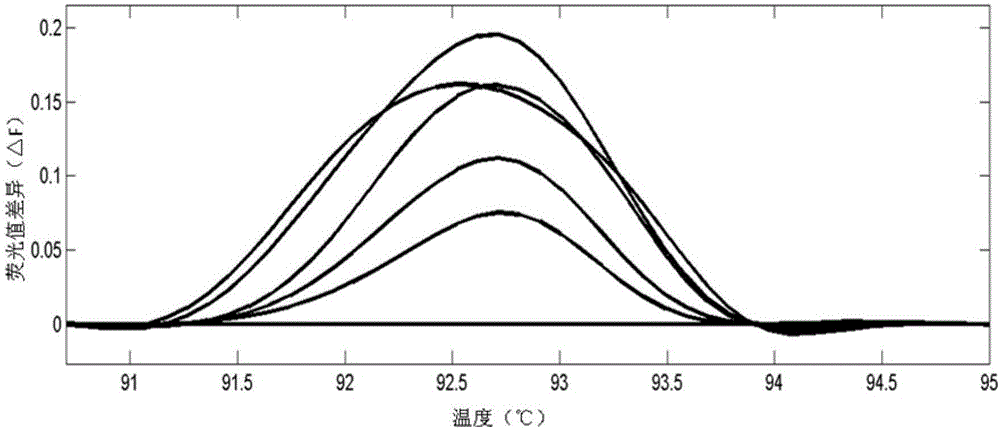
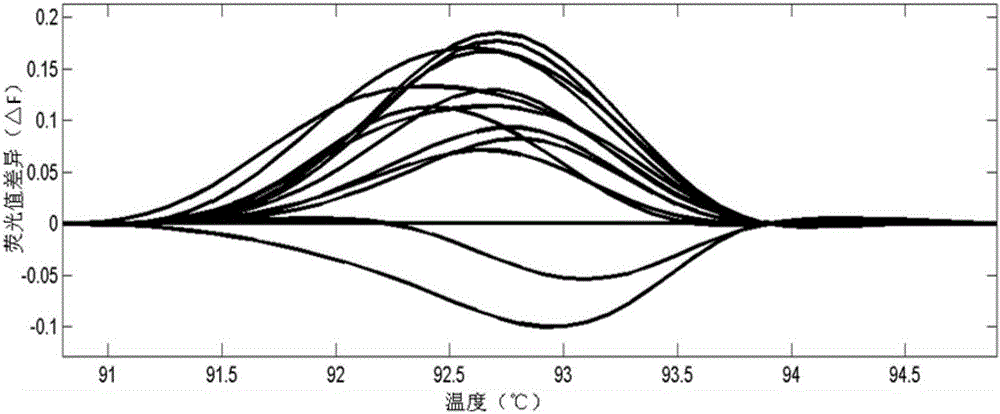
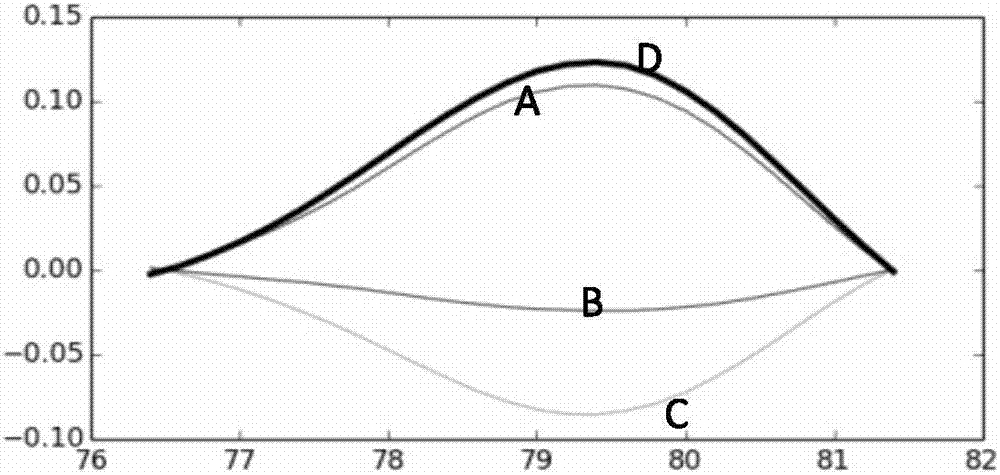
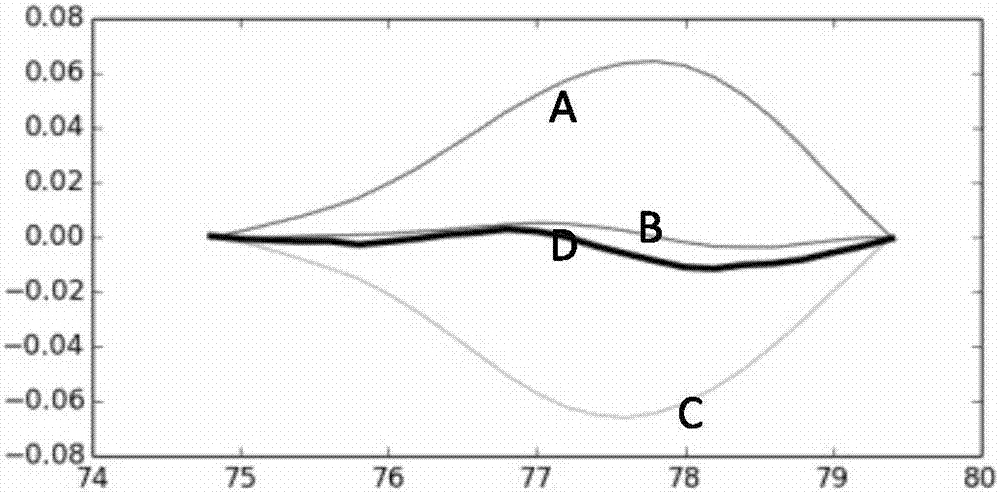
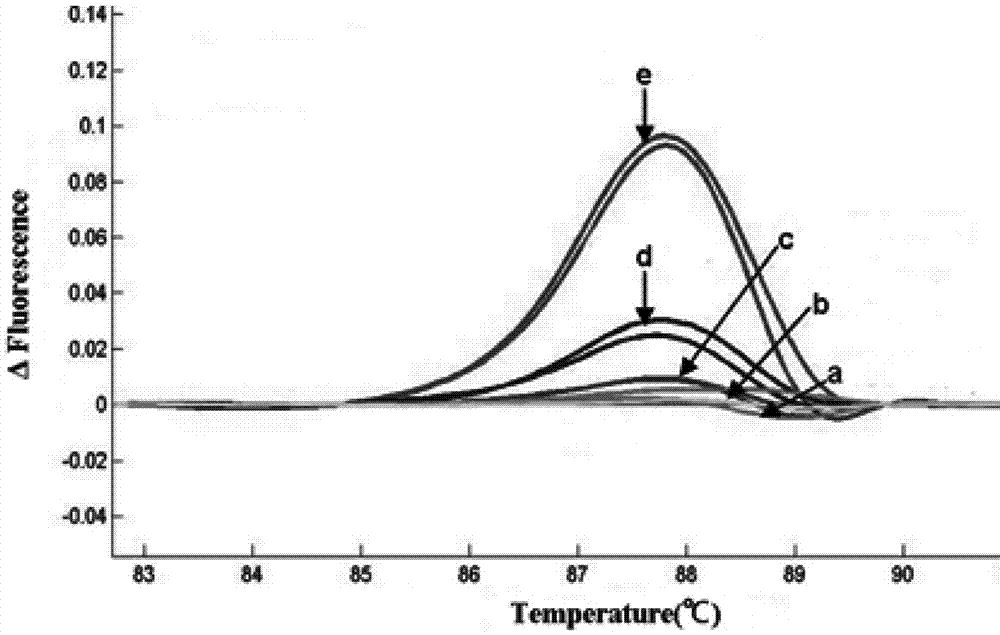
The invention discloses a method for screening a rice plant subjected to targeted gene editing. The method comprises the following steps: extracting genomic DNA of a rice plant to be detected and genomic DNA of a contrast rice plant, adding specific primers and a fluorescent dye, performing PCR amplification on DNA segments containing editing sites in the extracted genomic DNA, then, performing HRM (High Resolution Melting) analysis, and contrasting corresponding high resolution melting curves of the plant to be detected and the contrast plant by taking the high resolution melting curve corresponding to the contrast plant as a reference line to obtain the maximum fluorescence difference delta F; if the absolute value of the delta F is smaller than 0.05, determining that the plant to be detected is not successfully edited; if the absolute value of the delta F is not smaller than 0.05, determining that the plant to be detected is successfully edited. The method provided by the invention is simple, convenient, quick, effectively, accurate in result, high in flux, and low in use cost; when the method is used for assisting seed breeding, the plant screening efficiency can be greatly improved.
Owner:无锡哈勃生物种业技术研究院有限公司 +1
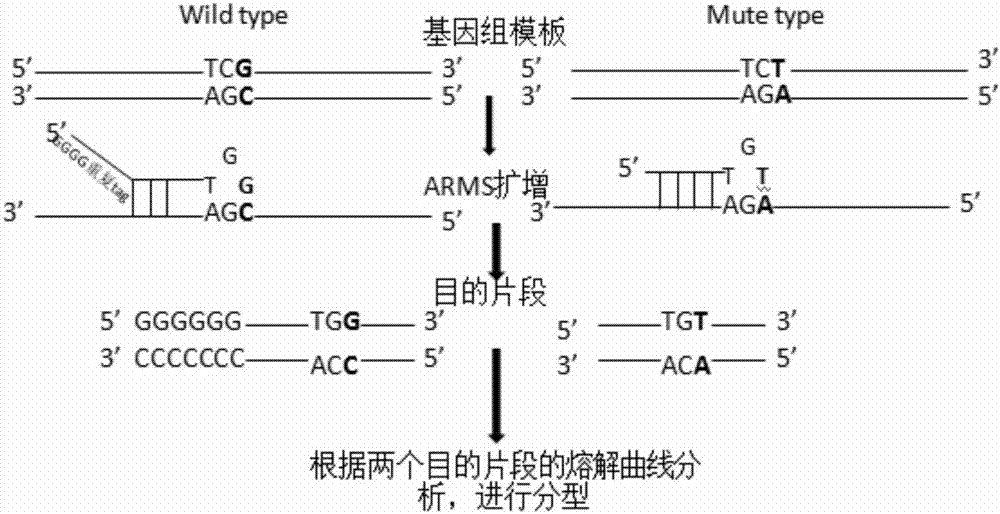
Human ApoE gene polymorphism detection kit based on ARMS-PCR melting curve method
PendingCN107460235AImprove the ability of SNP typingIncrease the difference in melting curvesMicrobiological testing/measurementFluorescenceTyping
The invention provides a human ApoE gene polymorphism detection kit based on the ARMS-PCR melting curve detection method. The human ApoE gene polymorphism detection kit comprises primers for detecting the polymorphism of an ApoE gene, and the primers include two groups of primers for detecting the c.388T>C site and the c.526C>T site respectively. The invention further provides an ARMS-PCR melting curve detection method and a PCR method of the human ApoE gene polymorphism detection kit. By the adoption of the human ApoE gene polymorphism detection kit based on the ARMS-PCR melting curve method, the ability of allele-specific primers to distinguish different alleles can be greatly improved, the difference between melting curves of two amplified allele fragments is greatly increased, the ability of SNP typing of the melting curve method is enhanced, melting curve method SNP typing can be achieved simply with an ordinary fluorescence PCR instrument instead of a high-precision high-resolution melting curve PCR instrument, the detection result is accurate, sensitivity is high, and accurate typing of a genomic DNA sample as low as 1 ng can be achieved.
Owner:SHANGHAI BIOMED UNION BIOTECHNOLOGY CO LTD
Primer, reagent box and method for detecting salmonella and integron (int)
InactiveCN104894283AReasonable compositionReasonable ratioMicrobiological testing/measurementAgainst vector-borne diseasesIntegrinCurve analysis
The invention discloses primer, a reagent box and a method for detecting salmonella and integrin (int). The primer comprises upstream primer SLMF: TGCGTCAGGACCTCATAG, downstream primer SLMR: ATGCCTGGAGGAGTGTTT, upstream primer intF: TTTACGCTGCTGTATGGT, and downstream primer intR for detecting int: TTATTGCTGGGATTAGGC. The reagent box is filled with 10 uL of HRM reaction premix liquid, 0.2-3.4 uL of primer, 1 uL of DNA template and is filled into 20 uL by water. The detecting method includes extracting sample DNA and performing PCR amplification. After amplification, products are subjected to high-resolution melting curve analysis, and results are judged by an amplification curve. The reagent box is reasonable in component and proportion, is convenient to use and quick and accurate to detect, the method is convenient and quick to operate, and low in cost, and detecting results are accurate.
Owner:蔡先全
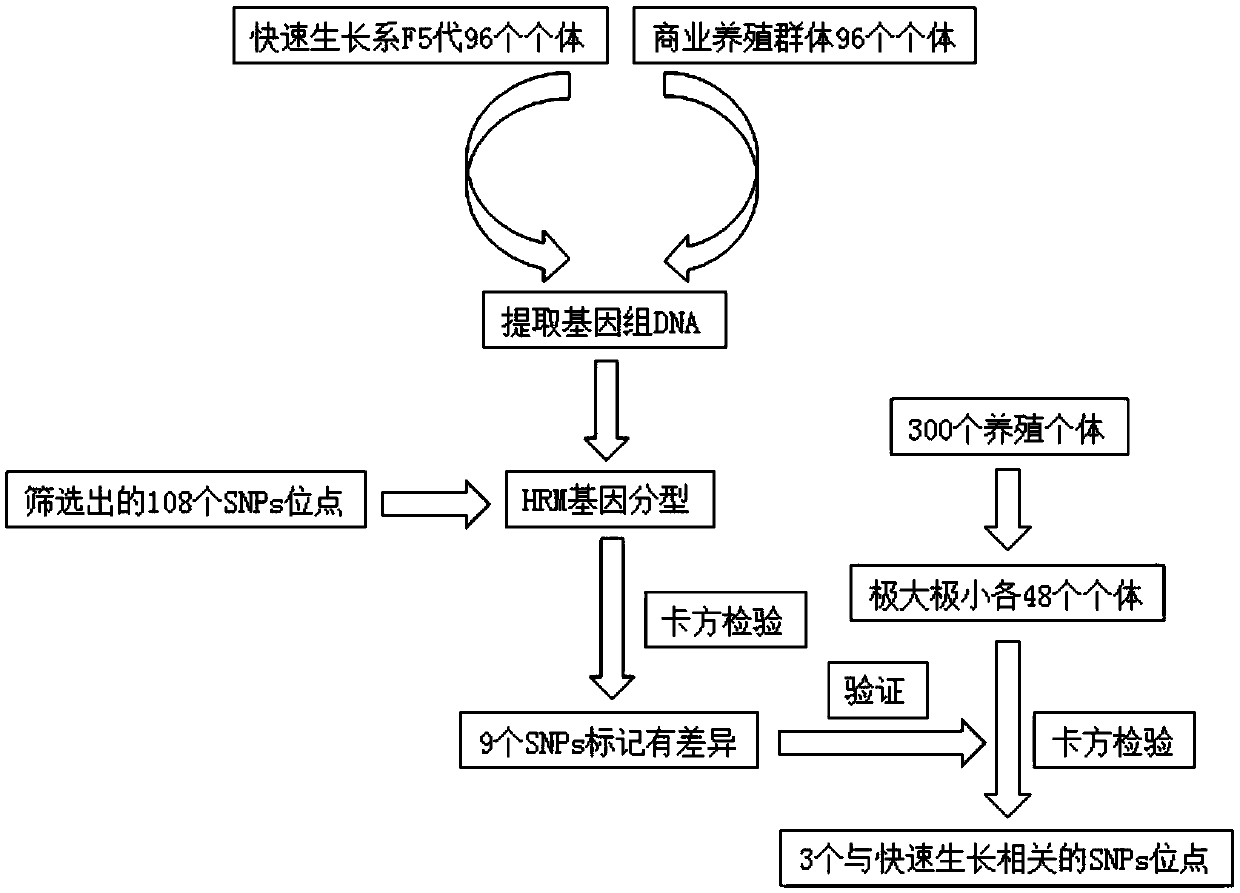
SNP marker related to rapid growth of pacific oysters and identification method as well as application thereof
ActiveCN105506162AImprove the efficiency of screening related genesGuaranteed reliabilityMicrobiological testing/measurementCorrelation analysisAllele
The invention discloses an SNP marker related to rapid growth of pacific oysters and an identification method as well as application thereof, and belongs to the field of molecular biological DNA marking technology and application. By applying a high-resolution melting curve analytic technique and utilizing 108 SNP loci mined from an EST common data base to perform genetic typing on pacific oyster rapid growth population after 5 generation breeding selection, 3 alleles closely related to the growth trait of pacific oysters are obtained via correlation analysis. The 3 SNP marker preponderant alleles can be applied to selection of rapid growth parent pacific oysters, and further provide a scientific auxiliary means for breeding selection of rapid growth pacific oysters. Compared with the conventional breeding method, the method has the advantages of strong purposefulness and direct action effect, besides, the operation is simple, the detection is rapid, the cost is low, and extensive popularization and application is facilitated.
Owner:OCEAN UNIV OF CHINA
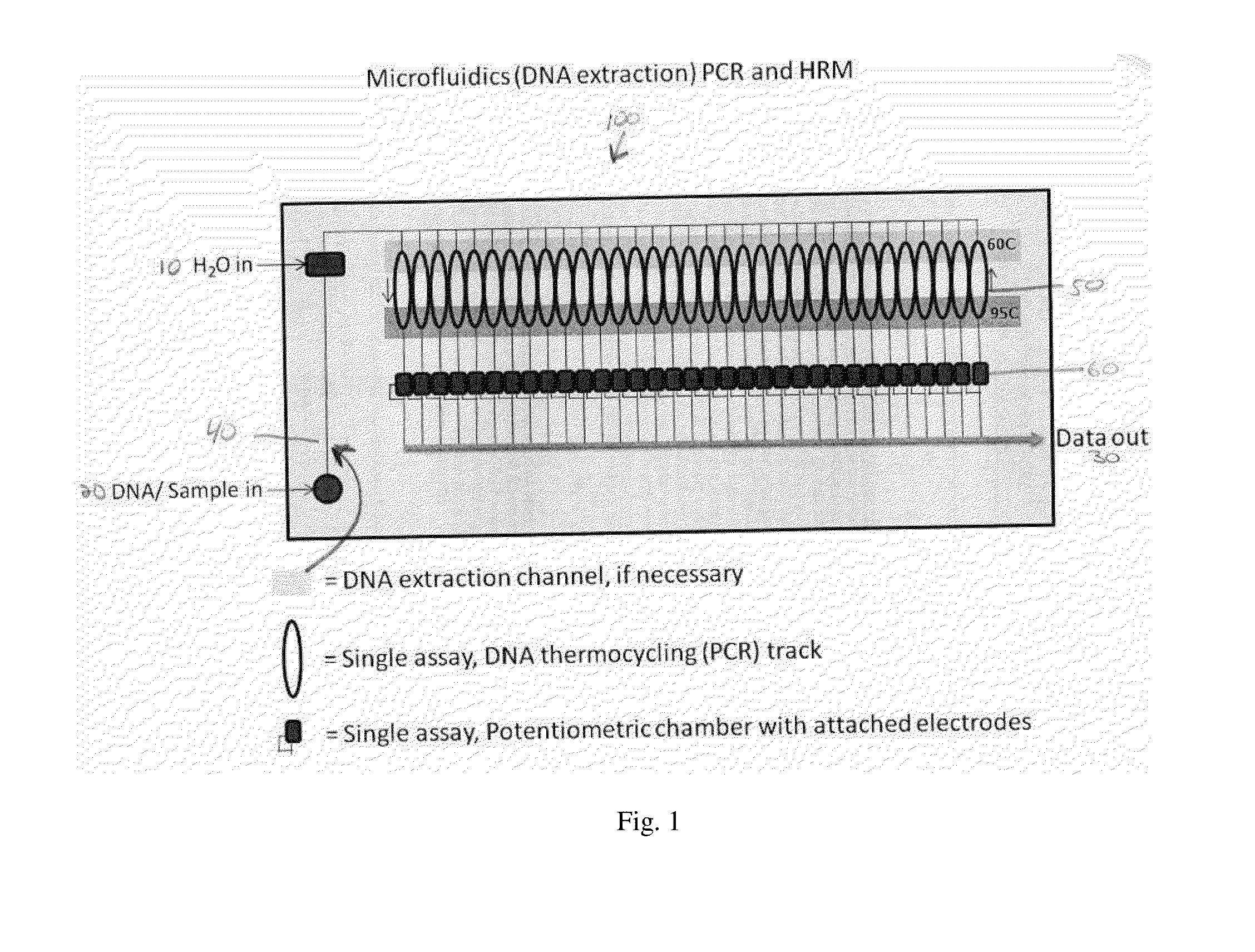
Microfluidics Polymerase Chain Reaction and High Resolution Melt Detection
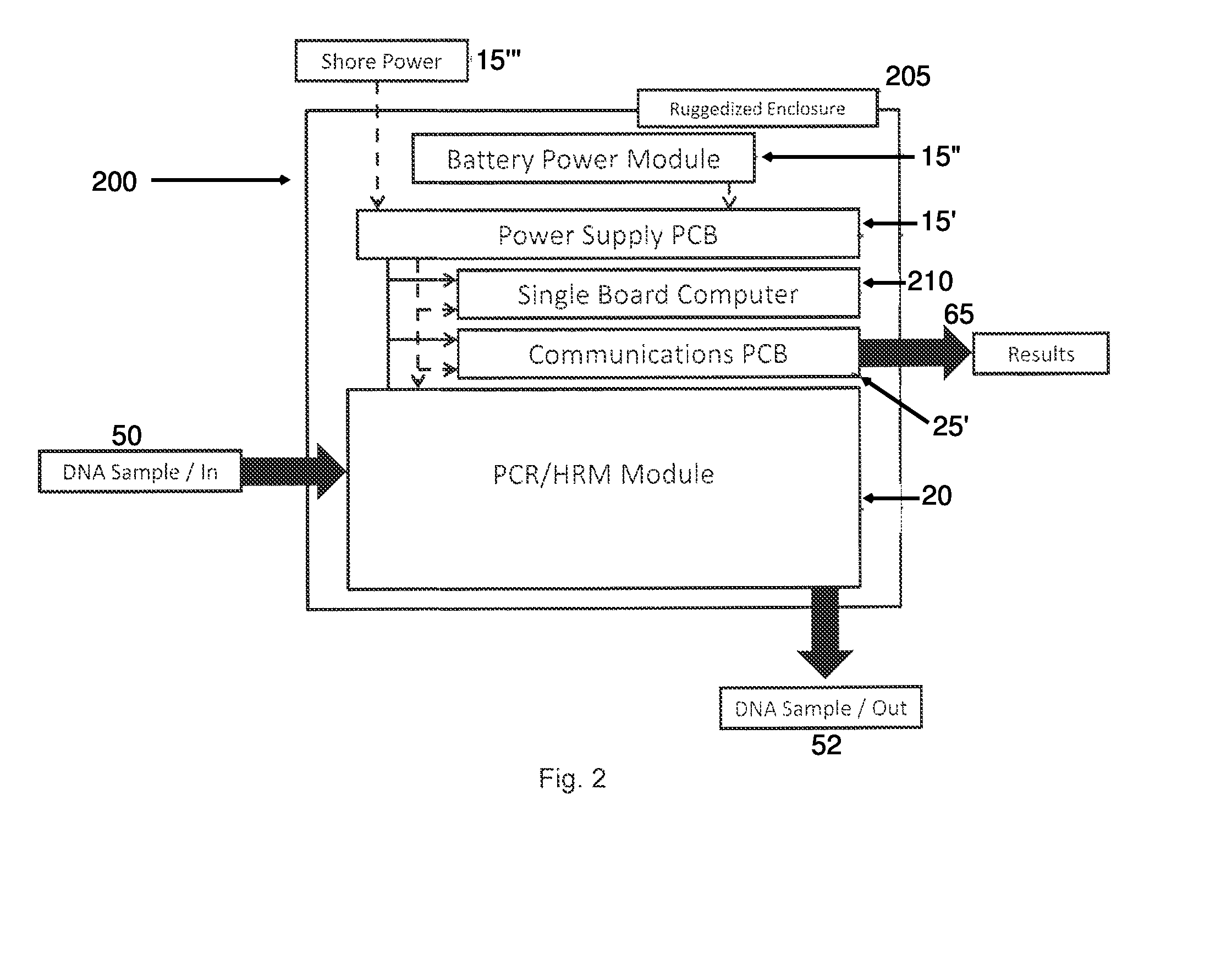
ActiveUS20140287420A1Rugged designBioreactor/fermenter combinationsBiological substance pretreatmentsBiologyHigh resolution
The present invention relates to a method and system for Polymerase Chain Reaction (“PCR”), High Resolution Melt (“HRM”) analysis and microfluidics, and, more specifically, to a method and system for implementing the processes of PCR and HRM on a microscale in a microfluidics chamber for certain purposes including for purposes of DNA detection and / or extraction.
Owner:SYRACUSE UNIVERSITY
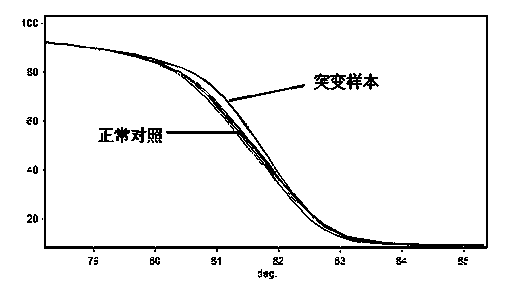
Method for detecting relative gene polymorphism of warfarin personalized medication by using high-resolution melting curve analysis technology
InactiveCN103525911AEasy to operateHigh sensitivityMicrobiological testing/measurementWarfarinSilica gel
The invention discloses a method for detecting relative gene polymorphism of warfarin personalized medication by using a high-resolution melting curve analysis technology. The method is characterized by comprising the following steps: adopting a silica gel adsorption method to extract genome DNA (Deoxyribose Nucleic Acid) of an oral epithelial cell / peripheral blood cell sample of a detected person; designing a detection primer containing a target site; carrying out site typing on a PCR (Polymerase Chain Reaction) amplified gene sequence by using the high-resolution melting curve (HRM) analysis technology. The method disclosed by the invention is high in sensitivity, good in specificity and fast in detection speed, and can be used for providing reference to a clinical medication dosage of warfarin, so as to realize reasonable and effective personalized medication.
Owner:上海中优医药高科技股份有限公司
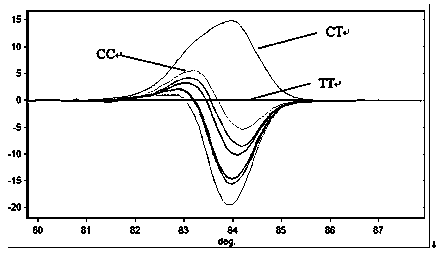
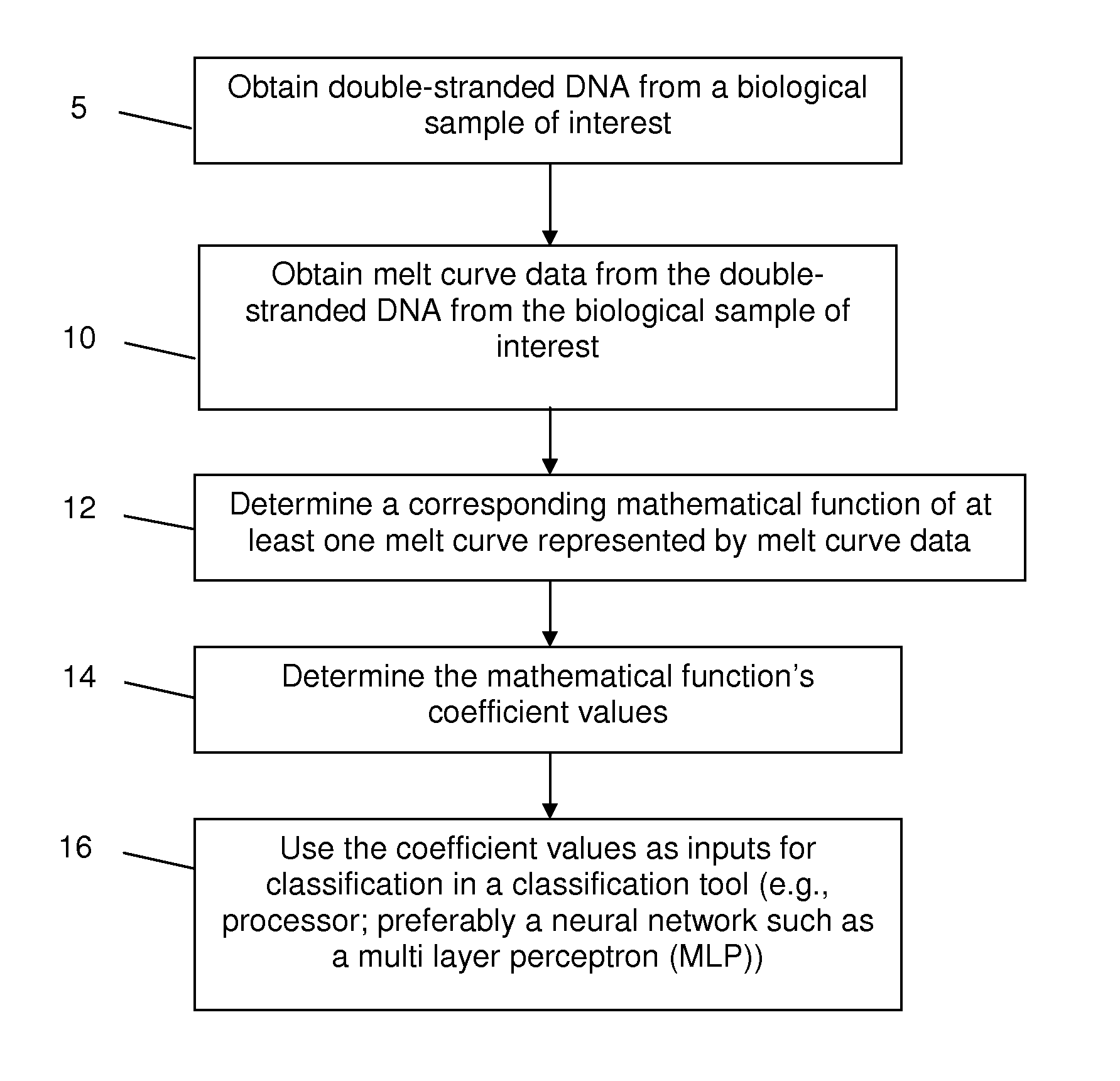
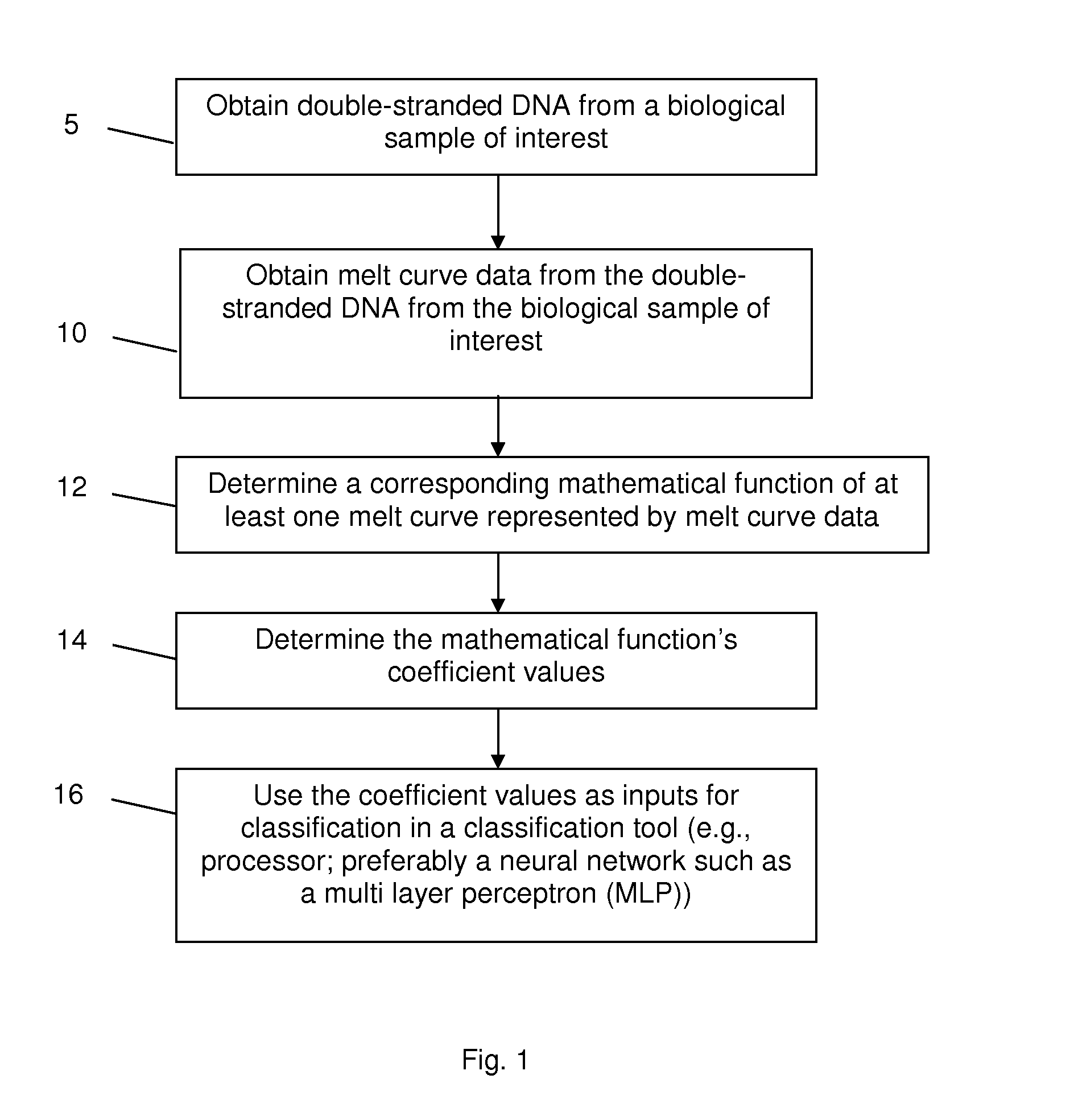
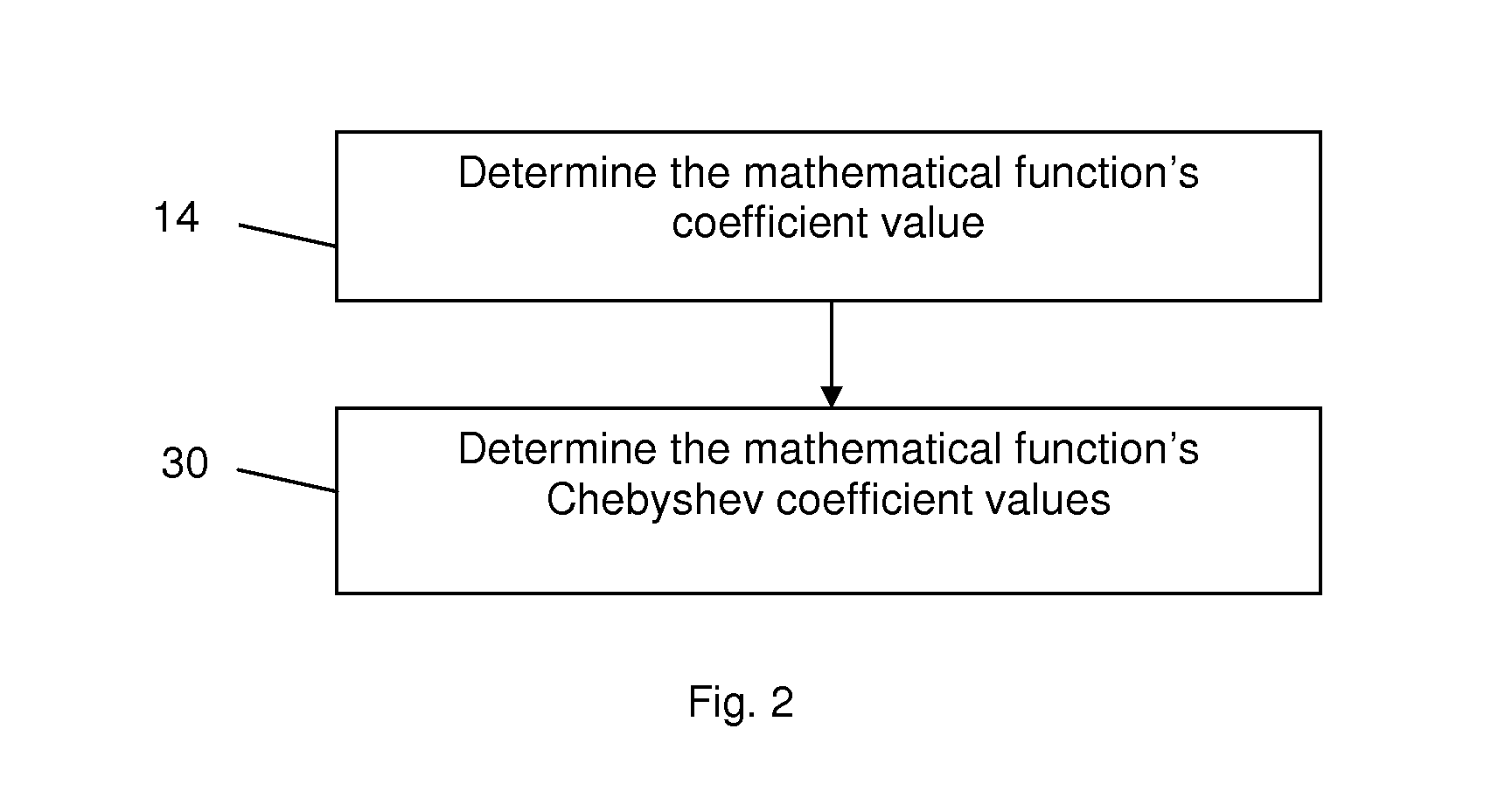
High-Resolution Melt Curve Classification Using Neural Networks
ActiveUS20140278126A1The result is accurateHigh resolutionBiostatisticsBiological testingAlgorithmMethod of undetermined coefficients
Owner:SYRACUSE UNIVERSITY
Method for detecting PAH (Polycyclic Aromatic Hydrocarbon) gene mutation by utilizing high-resolution melting curve analysis technique
InactiveCN103509865AQuick screeningFacilitate the establishment of early diagnosisMicrobiological testing/measurementPhenylalanine hydroxylaseExon
The invention discloses a method for detecting PAH (Polycyclic Aromatic Hydrocarbon) gene mutation by utilizing a high-resolution melting curve analysis technique. The method is characterized by comprising the following steps: adopting a silica gel adsorption method to extract the genome DNA of an oral epithelial cell / peripheral blood cell sample of a detected person, designing the third, sixth, seventh, eleventh and twelfth exon sequences of the PAH gene as detection primers of a template, and carrying out mutation detection on the gene sequences after PCR (Polymerase Chain Reaction) amplification according to the high-resolution melting curve analysis technique. The method has the advantages of high sensitivity, good specificity and high detection speed and can quickly screen common mutation sites of phenylalanine hydroxylase genes. The invention provides a new basis for the method for detecting PAH gene mutation.
Owner:上海中优医药高科技股份有限公司
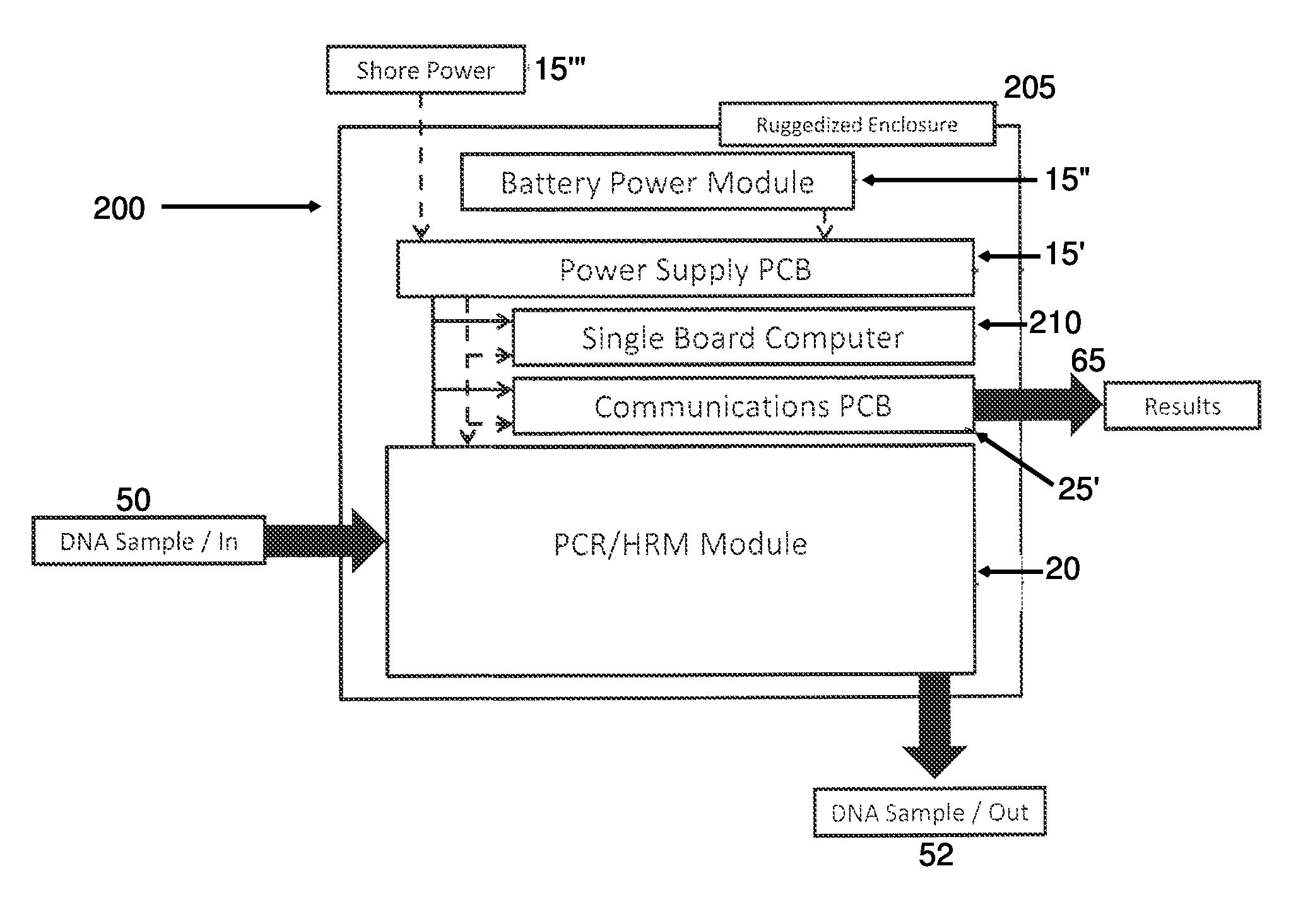
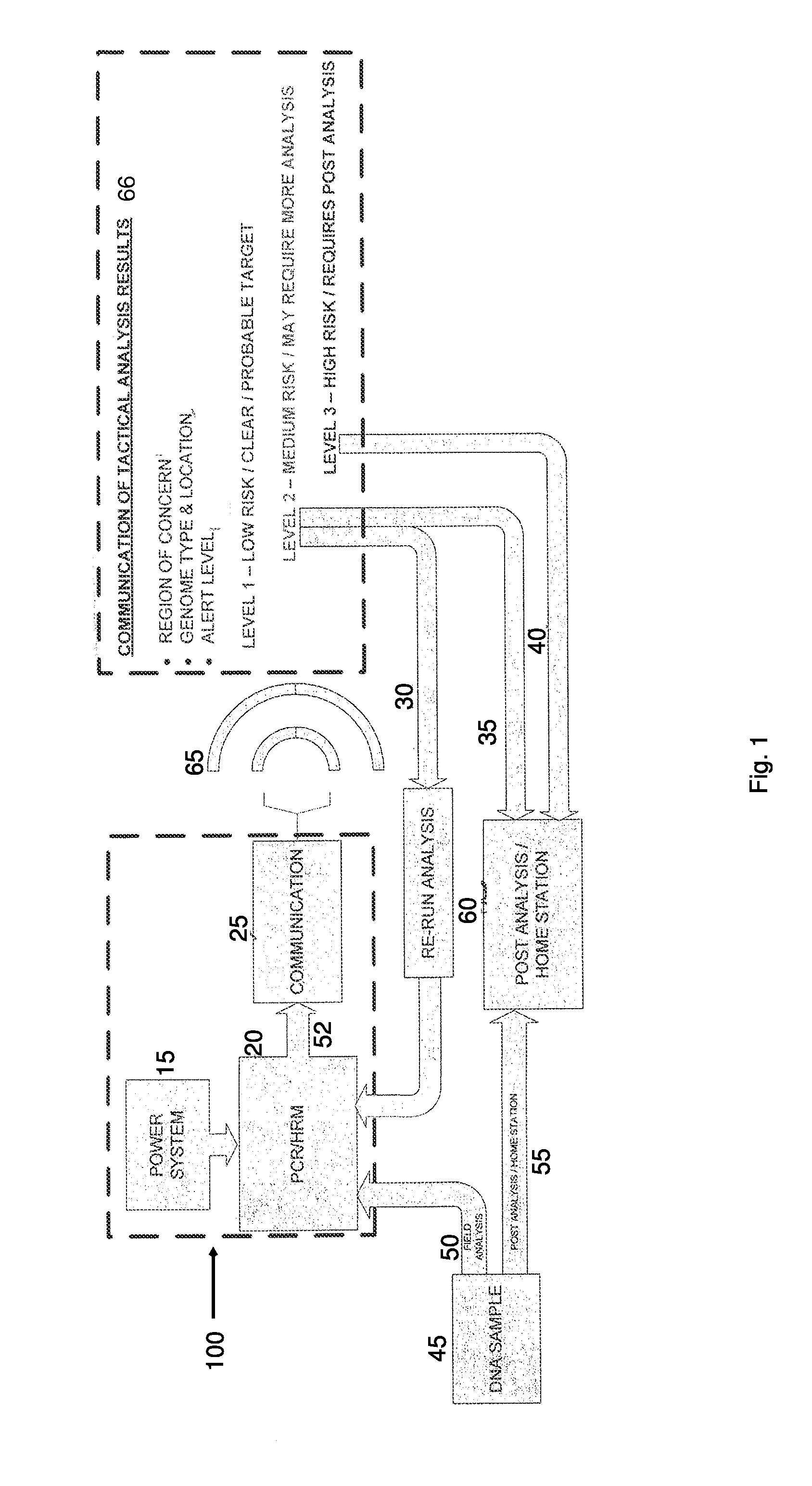
Tactical and portable PCR/HRM genome identification system
InactiveUS8956858B2Bioreactor/fermenter combinationsBiological substance pretreatmentsAlert levelBioinformatics
Owner:SRC INC
Single nucleotide polymorphism (SNP) molecular markers and method for pork DNA tracing by high resolution melting (HRM) method
ActiveCN104152447AHigh genetic polymorphismReduce testing costsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyAnalysis method
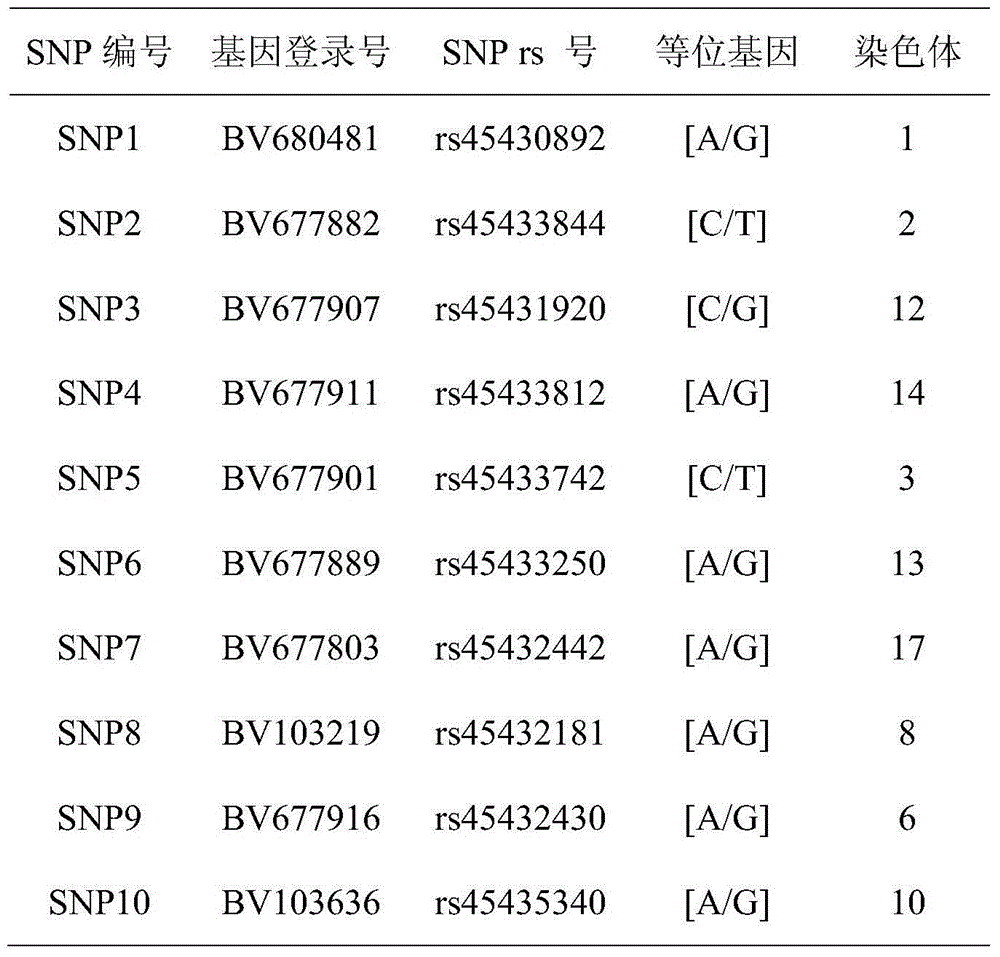
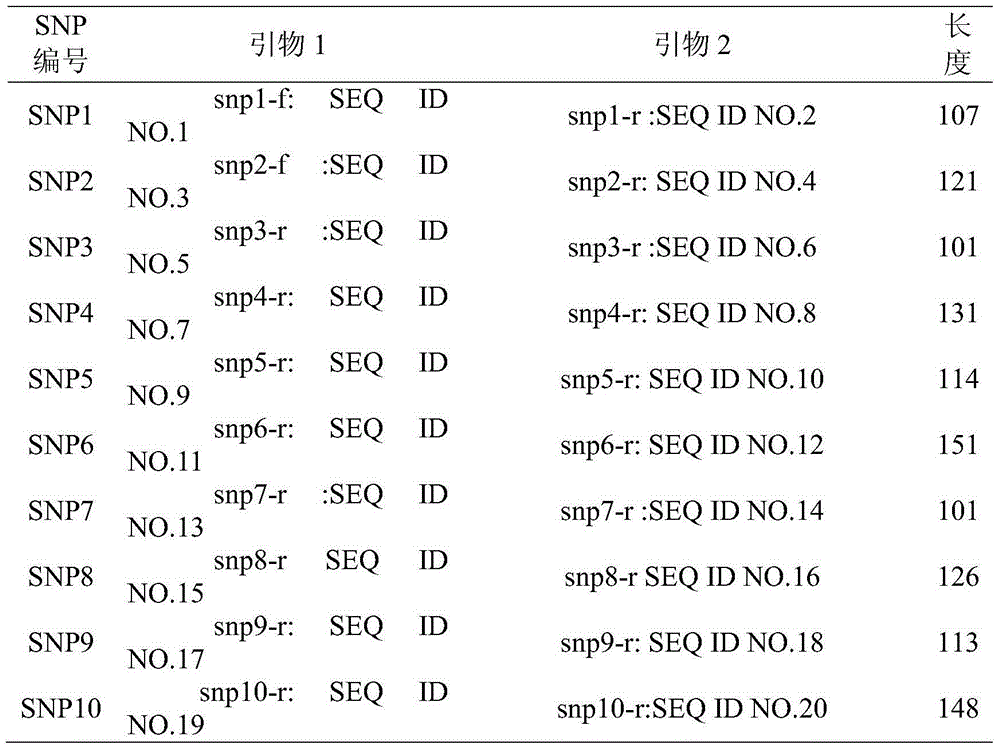
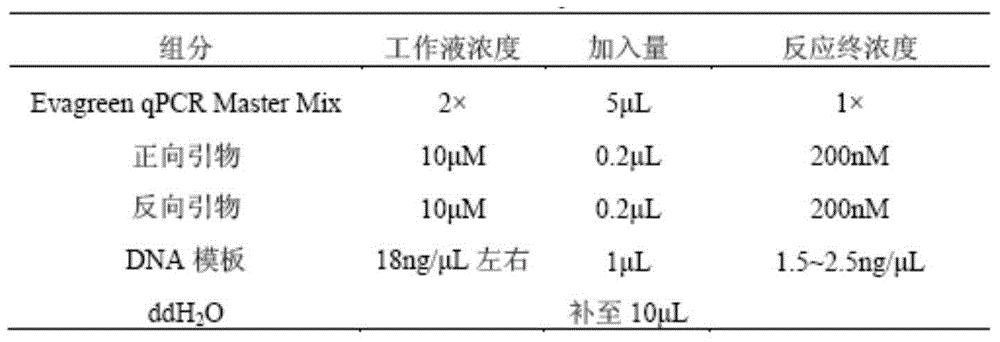
The invention belongs to the technical field of foods, provides single nucleotide polymorphism (SNP) molecular markers for pork DNA tracing and a method for pork DNA tracing by a high resolution melting (HRM) method. The SNP molecular markers comprise 10 SNPs molecular markers which are mutually independent, have gene heterogeneity at least 0.4 and are suitable as pork DNA tracing markers. The 10 SNPs molecular markers are obtained by a HRM analysis method, the effective SNPs sites form a pork SNPs information group and a pork DNA tracing process is carried out. The method for pork DNA tracing by the HRM method based on the SNP molecular markers has the characteristics of simpleness, speediness and low cost. The 10 SNPs molecular markers stably and widely exist in the pig genome DNA and are conducive to large-scale popularization of the DNA tracing technology.
Owner:NANJING AGRICULTURAL UNIVERSITY
Detection method of yak FOXO1 gene single nucleotide polymorphism and kit thereof
ActiveCN105441567APrecise positioningSensitive positioningMicrobiological testing/measurementNucleotideA-DNA
The invention relates to the technical field of gene polymorphism detection, in particular to a detection method of yak FOXO1 gene single nucleotide polymorphism and a kit thereof. The detection method comprises the following steps: 1) preparing a yak FOXO1 gene amplification product; 2) synthesizing a high and low temperature interior label; 3) preparing an HRM-PCR (High Resolution Melt-Polymerase Chain Reaction) amplification product; 4) collecting a fluorescence signal. Compared with an HRM method used for detecting polymorphic sites in the prior art, a gene probe designed by the detection method is more accurate and flexible in positioning, is accurate in parting, does not need PCR post-processing, has high working efficiency and good repeatability and is low in sample quality and quantity requirements, especially, the concentration requirement of a DNA template can be lowered to 0.1ng / mu L, and time and labor are saved when a sample size is large. Meanwhile, the detection kit has the advantages of high sensitiveness, high detection speed and good stability.
Owner:GANSU AGRI UNIV
Wheat spike density QTL linked HRM (high resolution melting) molecular marker and application thereof
ActiveCN107177667AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologySpike density
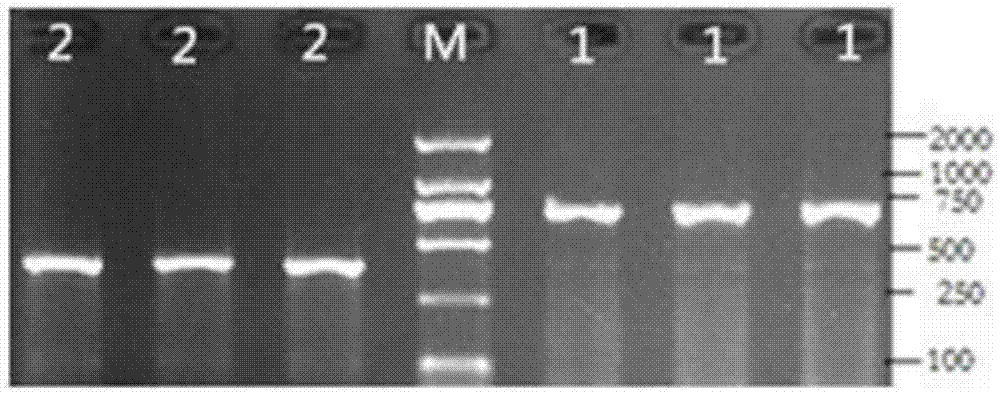
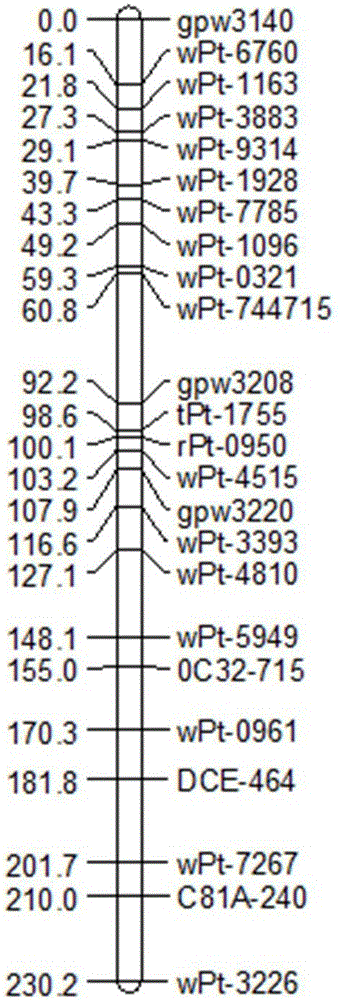
The invention discloses a wheat spike density QTL linked HRM (high resolution melting) molecular marker and application thereof and provides a wheat spike density QTL Qsd.sau-7A linked molecular marker. The molecular marker is 0C32-715, and a nucleotide sequence is shown as SEQ ID NO:1. According to detection and analysis, the molecular marker is capable of closely tracking wheat spike density QTL to predict spikelet dense insertion characteristics, and then selecting and breeding of a high-spike-density variety (strain) can be realized through molecular assisted selection. The invention further provides application of the molecular marker 0C32-715 to wheat breeding. By adoption of the method, accuracy in wheat spike density prediction can be improved in a certain range to make it convenient for quickly screening out the wheat variety or strain with spike density value increasing QTL for breeding, and accordingly a wheat high-yield variety selection process can be greatly accelerated.
Owner:SICHUAN AGRI UNIV
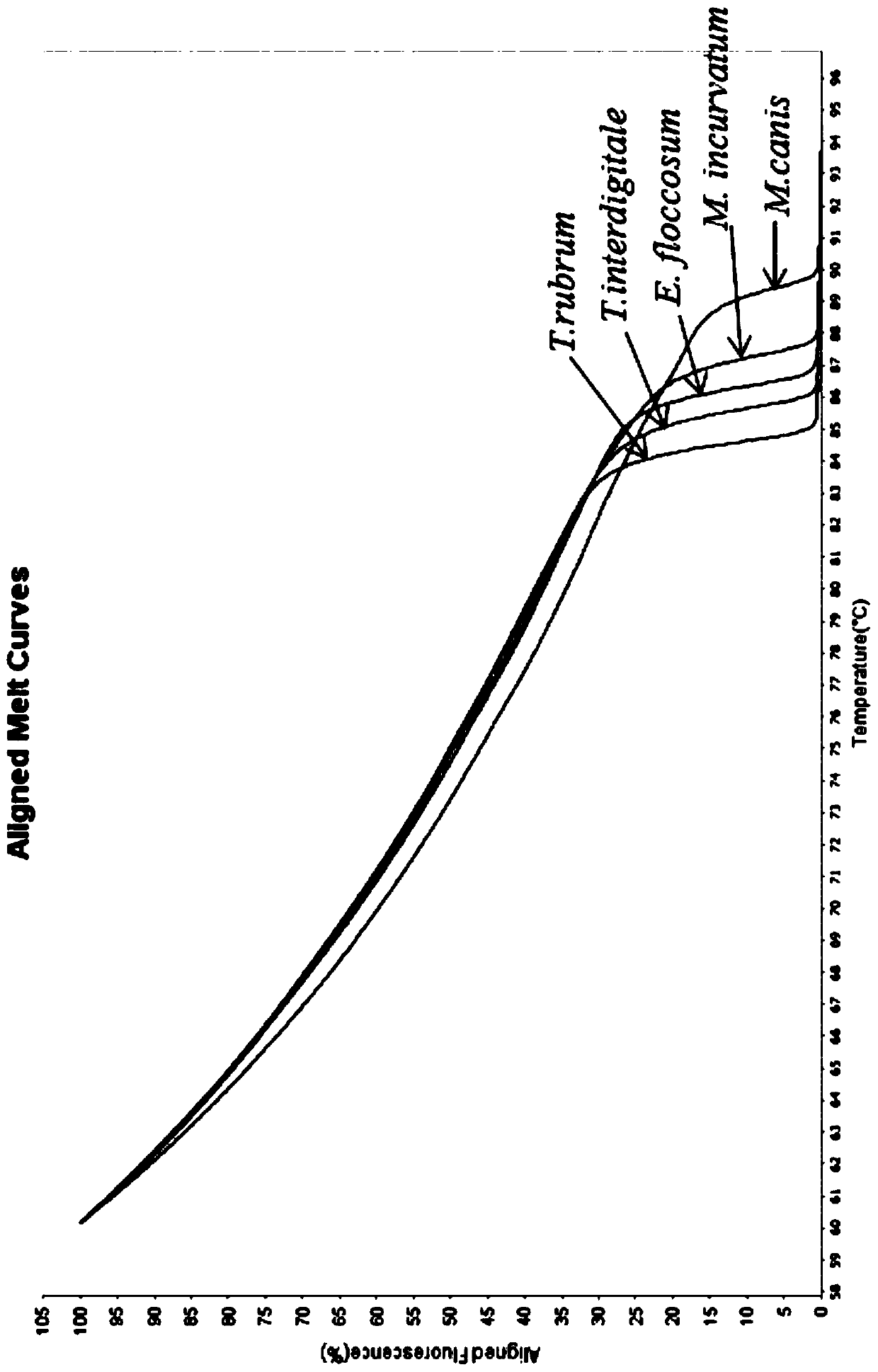
Method for identifying five dermatophytes by utilizing high-resolution melting curve
PendingCN110055347AAvoid cross contaminationEasy to operateMicrobiological testing/measurementMicroorganism based processesFluorescenceBiology
The invention provides a method for identifying five dermatophytes by utilizing a high-resolution melting curve and belongs to the technical field of high-resolution melting curve analysis. The methodadopts a pair of specific primers and is based on real-time fluorescence PCR melting curve to identify trichophyton rubrum, trichophyton digitorum, epidermophyton floccosum, microsporum caninum and Microsporum incurvatum. The sequence of the pair of specific primers is as shown as SEQ ID NO.1-2. The method has the advantages of high sensitivity, good specificity and high detection speed, can be used for identifying dermatophytes on the aspects of clinical diagnosis, environmental monitoring, food safety and the like, and provides a reliable basis for infection treatment, environmental sanitation and food safety monitoring of dermatophytes.
Owner:JINHUA MUNICIPAL CENT HOSPITAL
High resolution, high throughput hla genotyping by clonal sequencing
The invention provides methods and reagent for performing full, multi-locus HLA genotyping for multiple individuals in a single sequencing run using clonal sequencing.
Owner:F HOFFMANN LA ROCHE & CO AG
Method for detecting FOXH1 gene SNP locus rs750472 genotype
InactiveCN105441545AAccurate detectionHigh sensitivityMicrobiological testing/measurementGenomic DNAGenetics
The invention provides a method for detecting an FOXH1 gene SNP locus rs750472 genotype. The method comprises the following steps: (1) constructing a recombinant negative plasmid and a recombinant positive plasmid; (2) extracting a peripheral blood genomic DNA of a to-be-detected sample, carrying out PCR amplification and HRM analysis of the recombinant negative plasmid, the recombinant positive plasmid and the peripheral blood genomic DNA of the to-be-detected sample, and collecting data; and (3) drawing high resolution melting curves according to the collected data, and judging the FOXH1 gene SNP locus rs750472 genotype of the to-be-detected sample according to the melting curves of the recombinant negative plasmid and the recombinant positive plasmid. The method can effectively detect the FOXH1 gene SNP locus rs750472 genotype, and has the advantages of high sensitivity, good specificity, and rapid detection; and an analysis result with application of the method is entirely consistent to a sequencing result, and the method can be used for assistant judgment of ventricular septal defect heart disease.
Owner:SUZHOU BAIYUAN GENT CO LTD
Kit and method for detecting mutant alpha-Mediterranean anemia genes through HRM (high resolution melting) method
InactiveCN102925560AInhibitory complexEasy to detectMicrobiological testing/measurementFluorescence/phosphorescenceFluorescencePolymerase L
The invention relates to a disease gene detection technology, and particularly relates to a kit and method for detecting mutant alpha-Mediterranean anemia genes through an HRM (high resolution melting) method. The kit provided by the invention comprises two PCR (polymerase chain reaction) tubes, namely an alpha-PCR tube 1 and an alpha-PCR tube 2, wherein each PCR tube contains a PCR reagent; the PCR reagent comprises primers, a PCR Buffer, dNTPs (deoxyribonucleotide triphosphate), MgCl2, DNA (deoxyribonucleic acid) polymerase and saturated fluorescent dyes. By using the optimized PCR-HRM reaction conditions, 6 mutant alpha-Mediterranean anemia genes, including one of alpha-cod30, alpha-cod31, alpha-cod59, alpha-WM-cod122, alpha-QS-cod125 and alpha-CS-cod142 or a combination of more than one, can be simultaneously detected.
Owner:泰普生物科学(中国)有限公司
Detection primer for human IDH (isocitrate dehydrogenase) gene mutation and reagent kit
ActiveCN102732633ASimple and fast operationLow costMicrobiological testing/measurementDNA/RNA fragmentationHuman DNA sequencingBone marrow cell
The invention discloses a detection primer for human IDH (isocitrate dehydrogenase) gene mutation and a detection reagent kit, which are used for the combination of PCR (polymerase chain reaction) and high resolution melting curve analysis technique. The detection reagent kit can be used for detecting the gene mutation on IDH 1 and IDH2 gene exon 4, detection samples are human genomes DNA (deoxyribonucleic acid) extracted from tumor tissues, blood or bone marrow cells, and the detection reagent kit is used for patients with clinical brain glioma and hematopathy to assist tumor diagnosis, treatment and judging prognosis. The detection reagent kit is easy and rapid in operation and good in detection effect.
Owner:HELIXGEN GUANGZHOU
Transforming growth factor-beta (TGF-beta) type I receptor gene of chlamys farreri and single nucleotide polymorphism (SNP) locus of TGF-beta type I receptor gene
InactiveCN102899330AImprove seed selection efficiencyMicrobiological testing/measurementClimate change adaptationComplementary deoxyribonucleic acidMolecular genetics
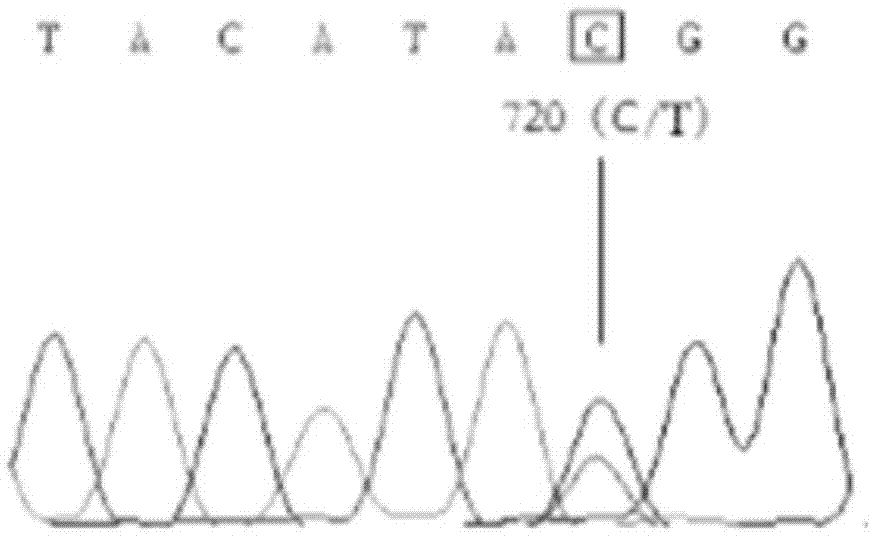
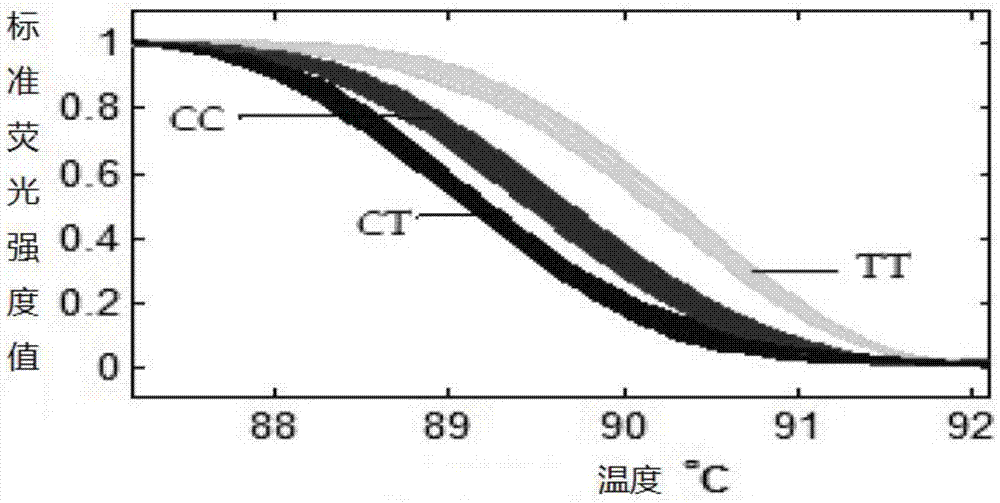
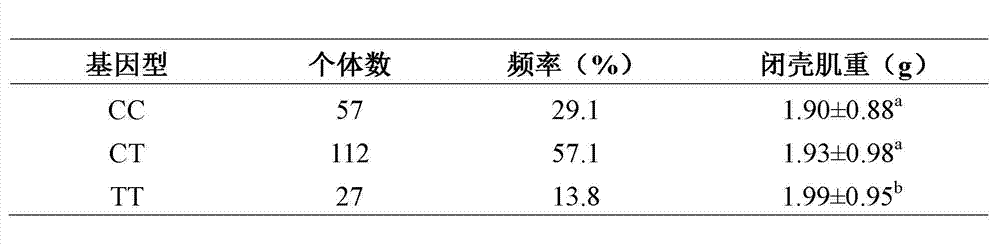
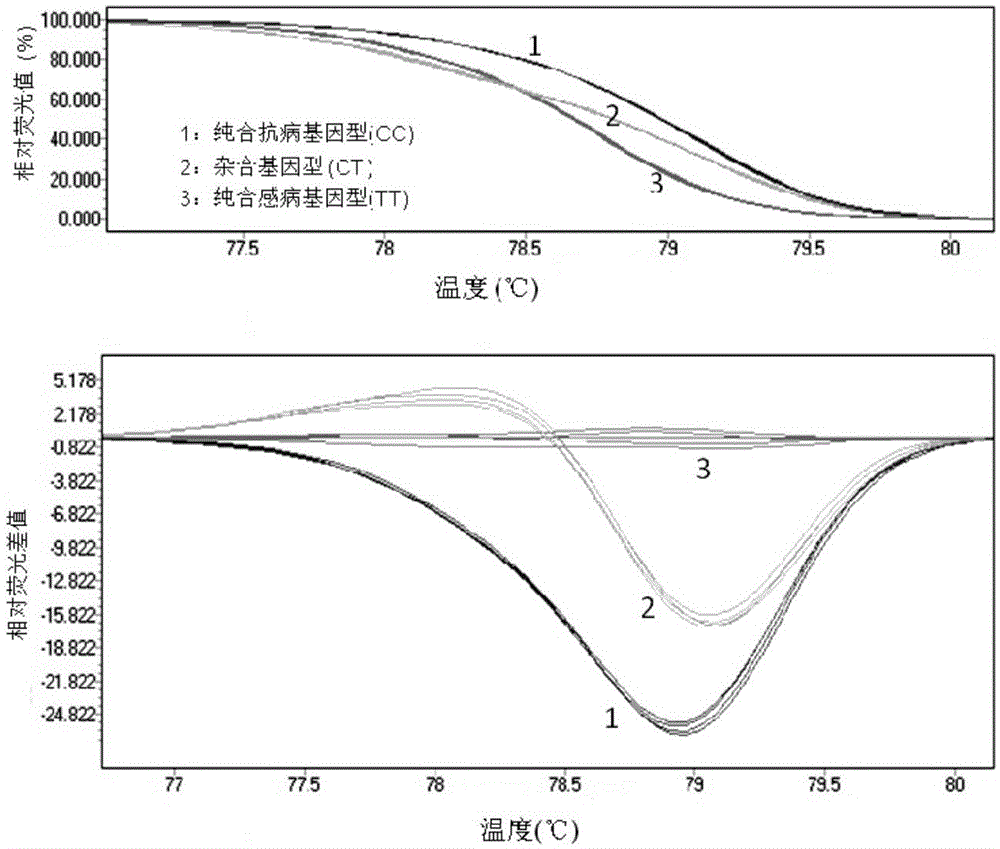
The invention relates to the cloning of a transforming growth factor-beta (TGF-beta) superfamily type I receptor gene Tgfbrl1 of chlamys farreri in a molecular genetic marker technology, a screening and typing technology of a single nucleotide polymorphism (SNP) locus which is relevant with the weight of adductor muscles in the gene and a method for the application of the gene to the breeding of the high-yield chlamys farreri. A total-length complementary deoxyribonucleic acid (cDNA) sequence of the Tgfbr1 gene of the chlamys farreri is obtained by utilizing a homology-based cloning strategy; an SNP lotus is discovered by blastn comparison, and primers and a probe are designed for the locus; and SNP typing is performed in a natural population of the chlamys farreri by using a high-resolution melting curve technology, and the weight of the adductor muscle of an individual is measured. Statistic analysis displays that the loci c.1815C>T are obviously relevant with the weight of the adductor muscles of the chlamys farreri, and the weight of the adductor muscles of individuals with TT genetypes is obviously higher than that of the adductor muscles of individuals with CC and CT genetypes. In the selective breeding process of the high-yield chlamys farreri, the breeding candidate colonies of the chlamys farreri can be subjected to c.1815C>T typing, and the individuals of which the loci c.1815C>T are TT genetypes are used as a breeding parent preferably by combining typing information of other loci which are relevant with growth properties.
Owner:OCEAN UNIV OF CHINA
High-resolution melting method and kit for identifying animal-derived components
ActiveCN109868322AImprove throughputGuaranteed repeatabilityMicrobiological testing/measurementMultiplex pcrsDNA
The invention discloses a high-resolution melting method and kit for identifying animal-derived components. The method comprises the following steps: extracting DNA in a sample as a template, and carrying out PCR amplification by adopting a universal primer; further melting an amplification product by adopting a high-resolution melting instrument; and analyzing the shape of a high resolution melting curve to identify animal derived components in the sample. According to the invention, the target gene sequence is amplified by adopting the universal primer, so that not only can the animal-derived components in a sample be accurately identified, but also the problems of competition between primer pairs, high PCR reaction condition optimization difficulty and the like are overcome compared with the traditional multiplex PCR, and the detection efficiency is obviously improved. The method has the advantages of simplicity in operation, high detection speed, high detection sample flux and accurate detection result, and is suitable for adulteration detection and traceability identification of animal-derived components in dairy products and other foods.
Owner:SHAANXI UNIV OF SCI & TECH
Method for genetic testing in single cells by HRM (high resolution melting) and pyrosequencing
InactiveCN106636435APrevent diseaseImprove population qualityMicrobiological testing/measurementDisease familyMelting curve analysis
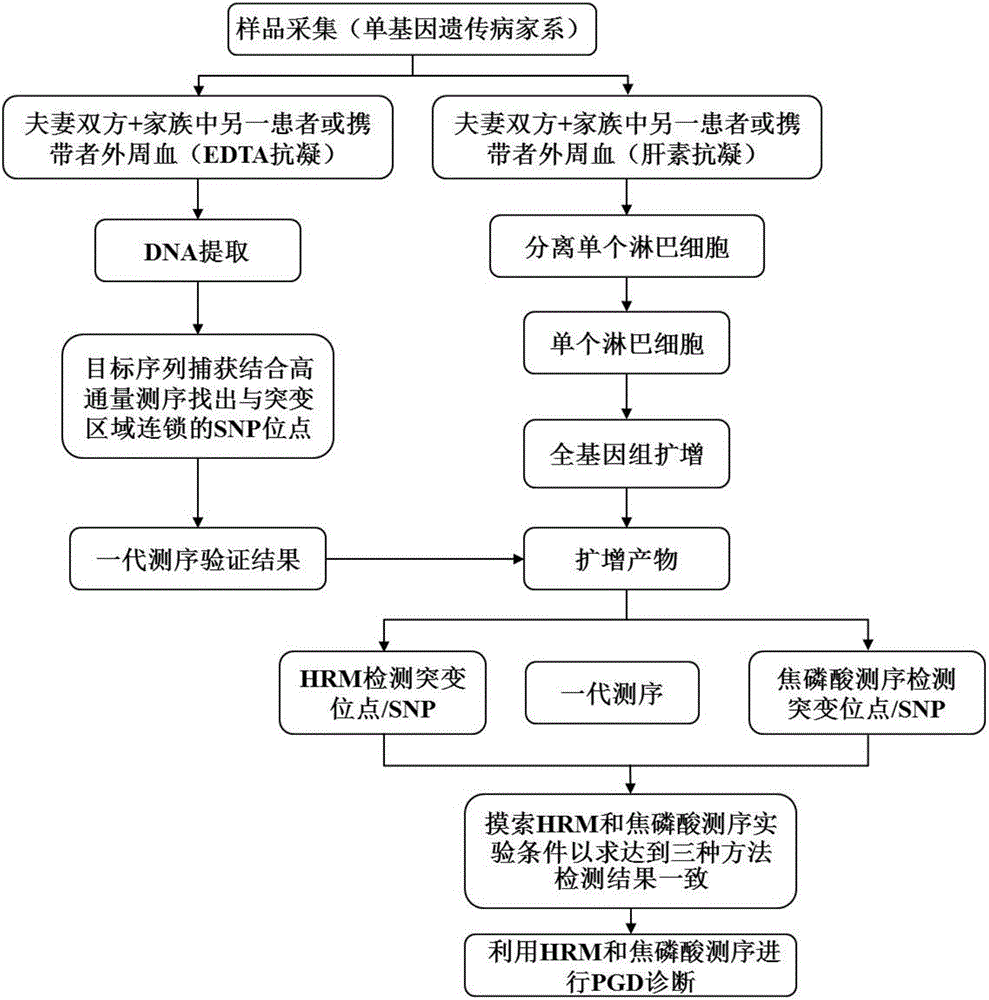
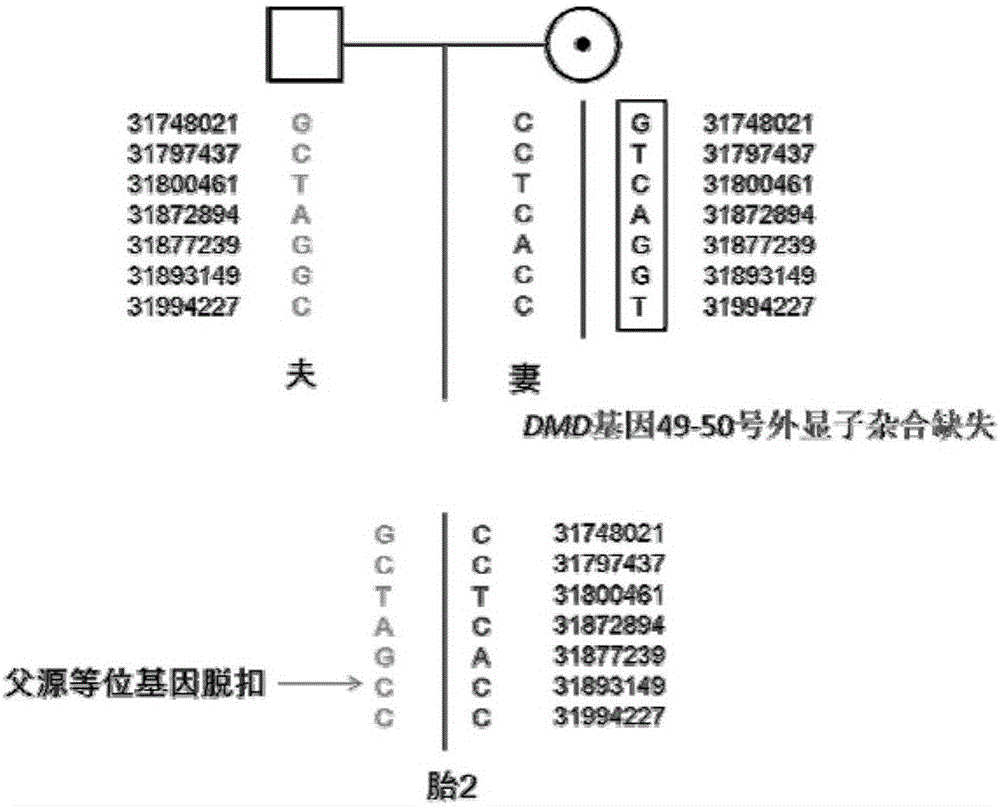
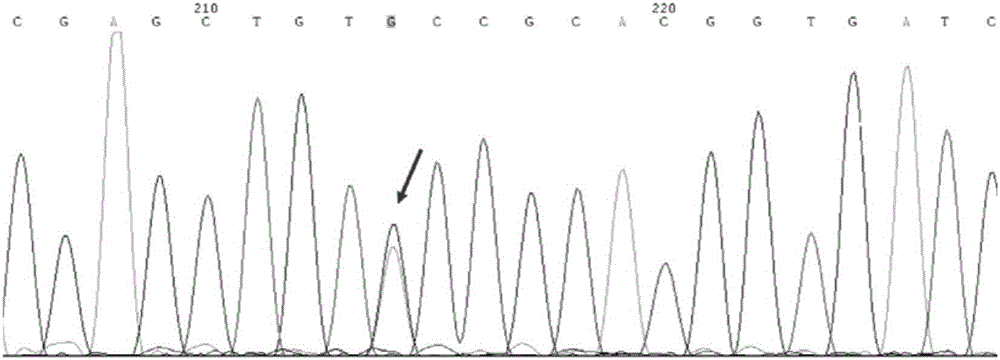
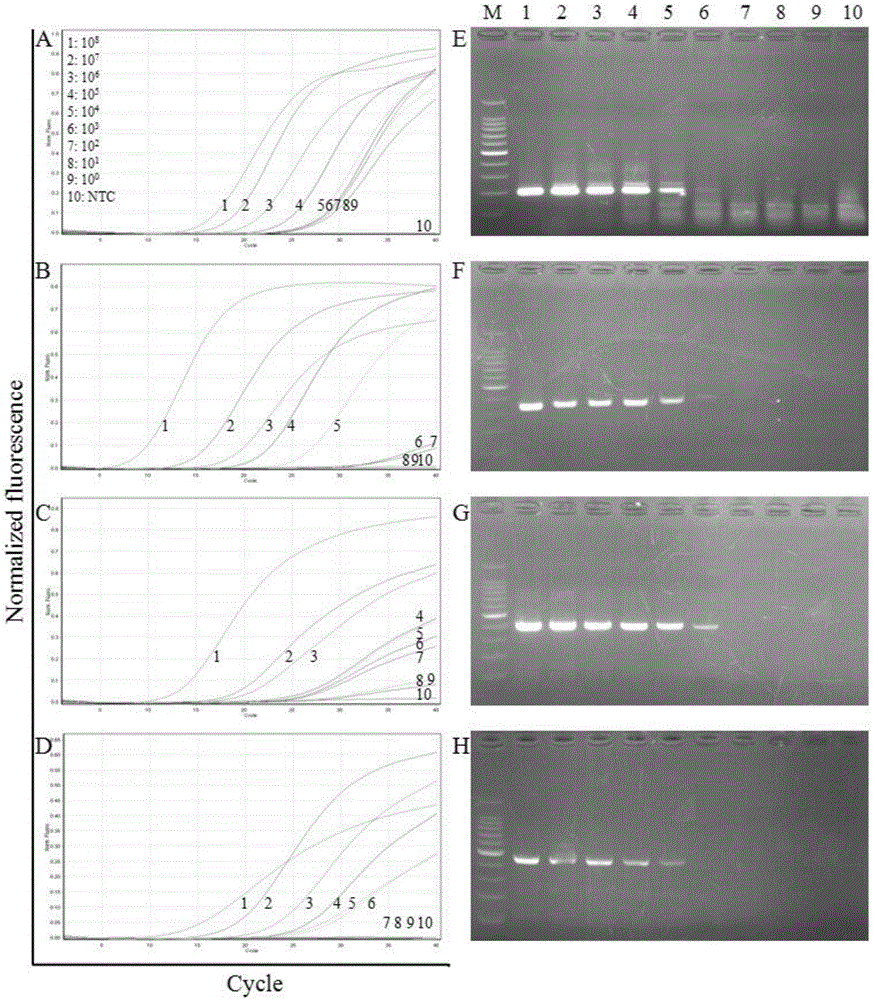
The invention belongs to the technical field of medical science and discloses a method for genetic testing in single cells by HRM (high resolution melting) and pyrosequencing. By high-throughput sequencing for haplotype analysis of a single-gene inheritance disease family, an analysis method for mutation detection and SNP (single nucleotide polymorphism) by adoption of a single-cell whole genome amplification product as a template on the basis of HRM and pyrosequencing is established. The method can be applied clinically directly to provide fast detection means for PGD (preimplantation genetic diagnosis) diagnosis of patients suffering from single gene inheritance diseases.
Owner:ZHEJIANG UNIV
Primer group and kit for simultaneously detecting four diarrhea viruses
InactiveCN104651538AEasy to detectHigh sensitivityMicrobiological testing/measurementMicroorganism based processesRotavirus RNAConserved sequence
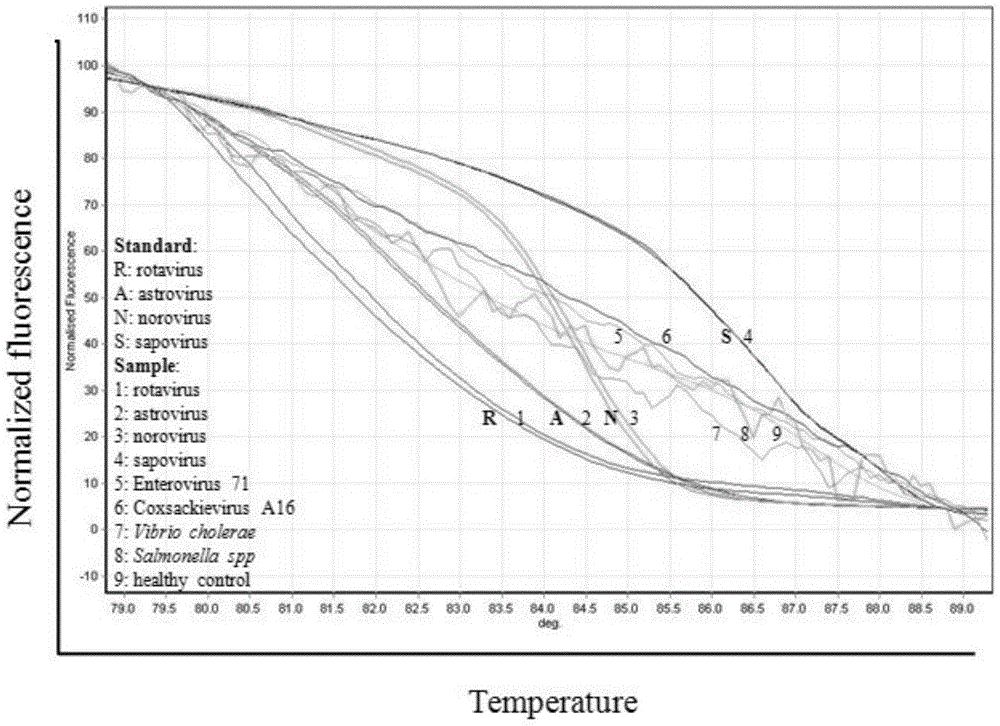
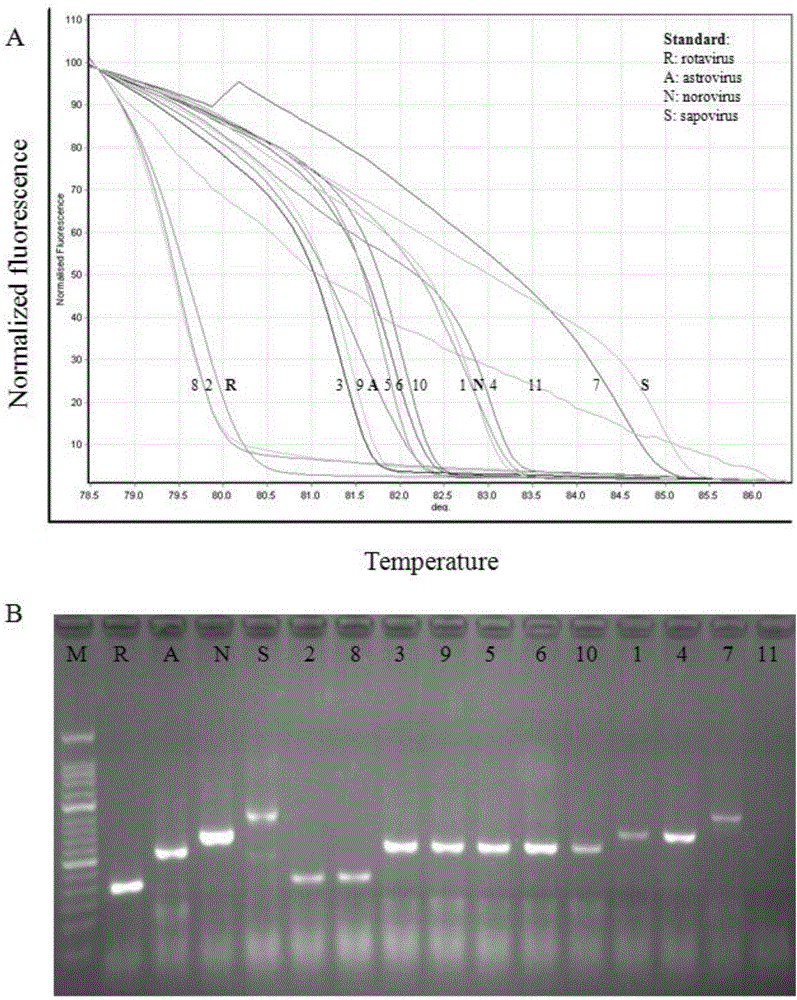
The invention discloses a primer group which is based on a high-resolution melting curve analysis technique and used for simultaneously detecting four diarrhea viruses, and a quadruple detection kit containing primers. The kit is capable of specifically detecting four diarrhea viruses, that is, rotavirus, astrovirus, norovirus and sapovirus. Through comparative analysis on respective homologies of the four viral genomes, respective conserved sequence design primers with high homologies can be found, the four diarrhea viruses can be simultaneously detected by using the high-resolution melting curve analysis technique, the primer group has the characteristics of being simple, convenient, rapid, cheap, safe, high in sensitivity and high in specificity, and powerful technical support can be provided for prevention and control and clinical diagnosis of dysentery.
Owner:珠海国际旅行卫生保健中心
Molecular marker subjected to co-segregation with pea powdery mildew resistance allele er1-6 and application thereof
InactiveCN105039336AImprove throughputSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationCandidate Gene Association StudyBiology
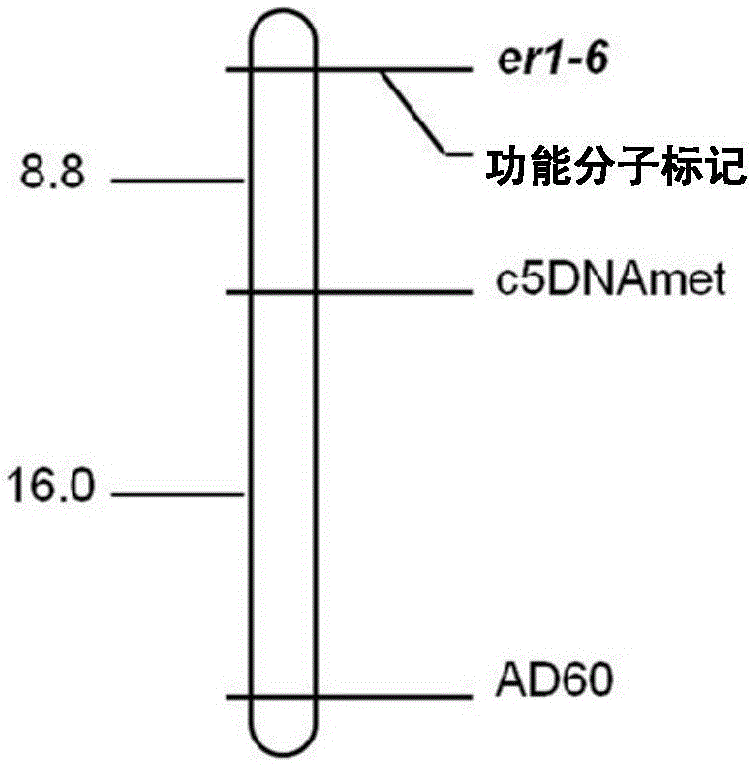
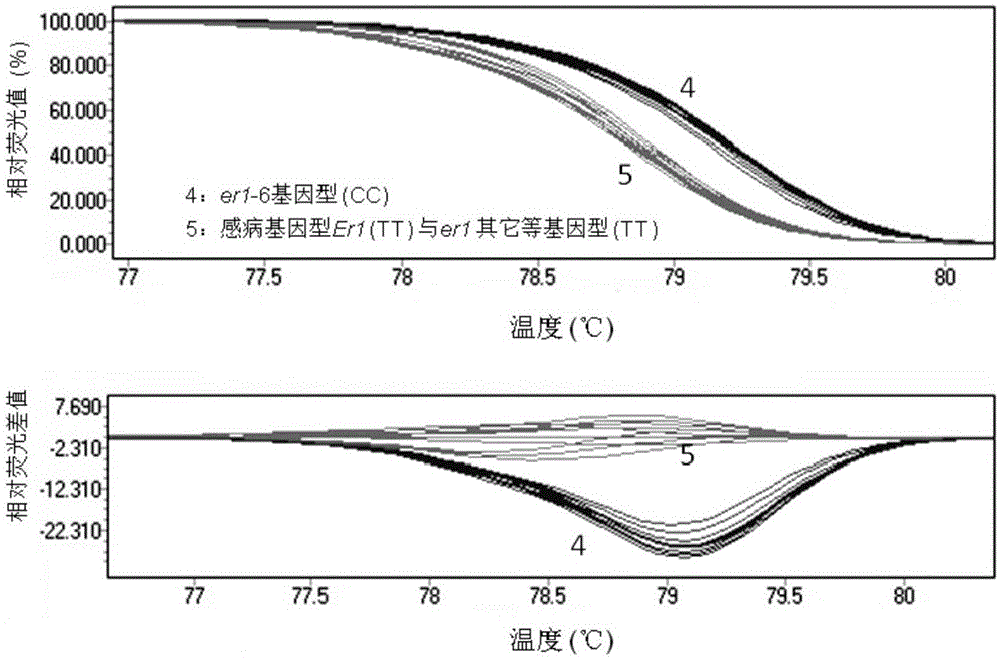
The invention provides a molecular marker subjected to co-segregation with a pea powdery mildew resistance allele er1-6. The molecular marker is located on a VI linkage group of a pea genetic map, and the genetic distance between the molecular marker and the allele er1-6 is 0cM. According to the single base difference (at the position of 1121, T-C) between a disease-resistant variety G0001778 containing the resistance allele er1-6 and an er1 candidate gene PsMLO1cDNA sequence of infected variety pea number 6, primers are designed on the two sides of an SNP mutation site, a high resolution melting curve analysis technology is used for developing the molecular marker subjected to co-segregation with the pea powdery mildew resistance allele er1-6, the marker is subjected to group detection of F2 and F3 which are derived by the disease-resistant variety G0001778 and the infected variety pea number 6 in a hybridization mode, and it is verified that the marker is a co-dominance functional marker subjected to co-segregation with the gene er1-6. Afterwards, the effectiveness is verified in pea powdery mildew resistance resource identification through the marker, the functional marker can be used for molecular marker-assisted selection breeding of the pea powdery mildew resistance, and therefore the breeding process is accelerated.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com