Preparation method of non-transgenic CRISPR mutant
A non-transgenic, mutant technology, applied in the field of preparation of non-transgenic CRISPR mutants, can solve problems such as inability to pass phenotypic judgment, inability to use antibiotic screening, inability to visually determine phenotypic changes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
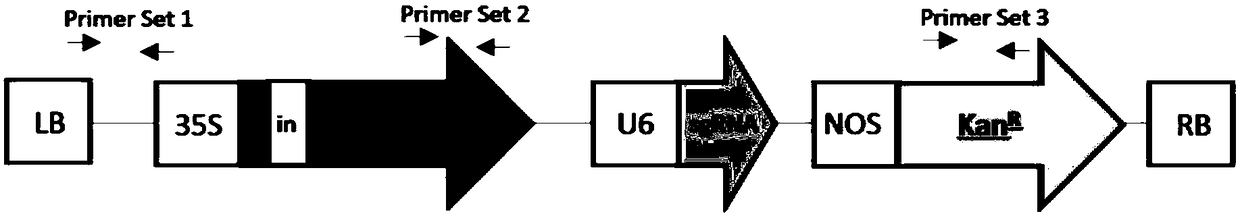
[0060] Embodiment 1. Construction of Cas9 mutation carrier
[0061] 1. Carrier sequence with 15 bases at the 5' end of the 5'-SpeI restriction site-SpeI restriction site-Cas9 gene sequence containing 1 intron-ApaI restriction site-ApaI restriction site Point the 15-base carrier sequence at the 5' end-the sequence at the 3' end, and directly synthesize a Cas9 gene (SEQ ID NO.1) containing a complete intron. Using restriction endonucleases SpeI and ApaI to double digest and linearize the CRISPR base vector pK7WGF2::hCas9 (Addgene plasmid #46965) at 37°C overnight, use the Infusion method to connect the Cas9 gene containing introns into the vector to obtain Intronic Cas9 vector. 20 μl common PCR amplification and sequencing to verify whether the vector connection is correct. The PCR reaction system is shown in Table 1. The PCR reaction conditions are: 94°C for 30s, 55°C for 30s, 72°C for 1min, repeating 30 cycles.
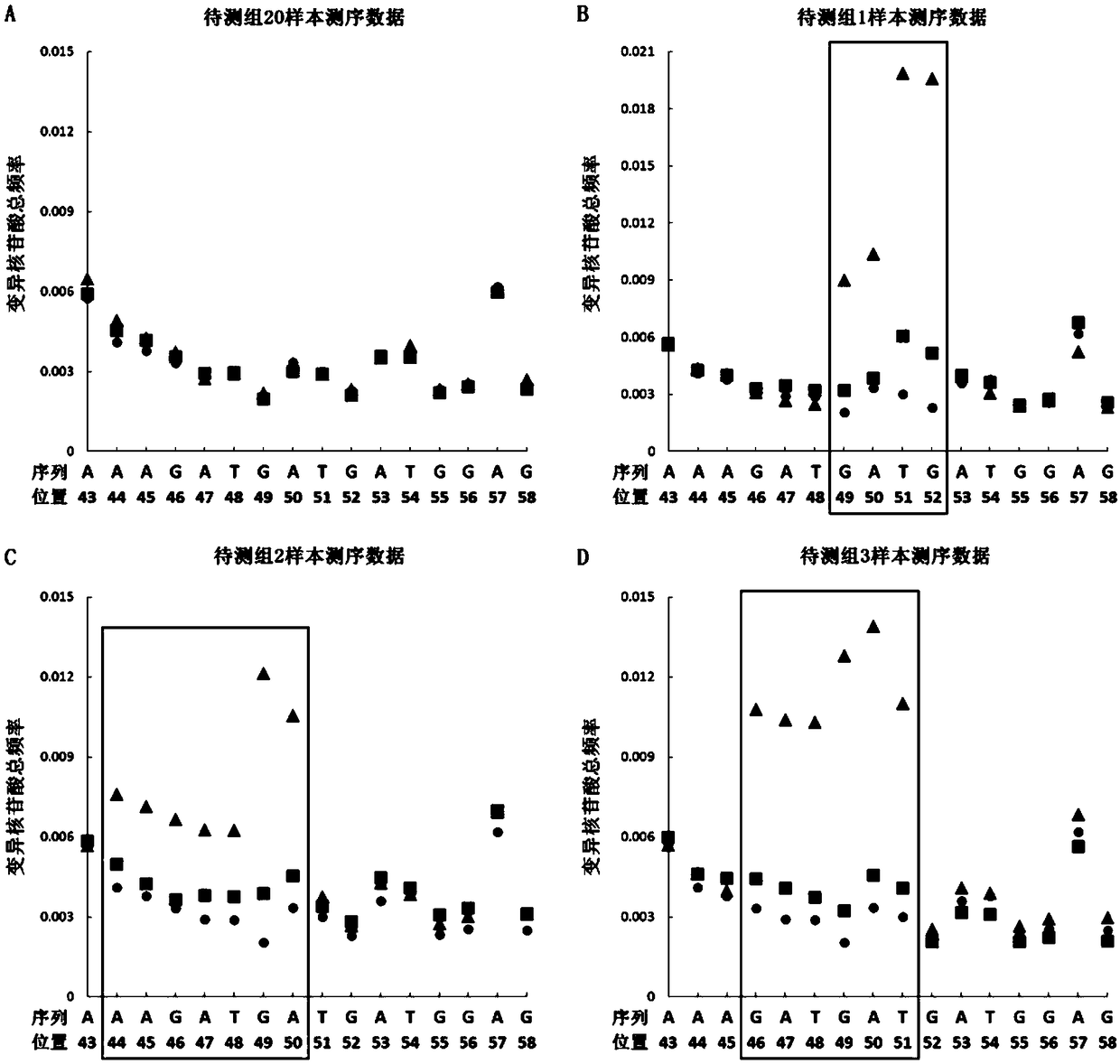
[0062] 2. According to the sequence of the fourth exon of th...
Embodiment 2
[0068] Example 2. Transformation of Agrobacterium
[0069] The PDS-Cas9 mutant vector was transformed into Agrobacterium EHA105 competent cells by freeze-thaw method. The specific steps are as follows: Add 0.1-1 μg (5-10 μl) of plasmid DNA to 50 μl of Agrobacterium competent cells, and ice-bath for 30 minutes; put it in liquid nitrogen for 5 minutes (or 1 minute), and then immediately put it in a 37°C water bath for 5 minutes ;Take out the centrifuge tube, put it on ice for 2min immediately, then add 900μl of LB without any antibiotics, shake and culture at 28℃, 220rpm for 3-5h; take out the bacterial liquid, centrifuge at 3000 rpm for 3min, then remove 850μl of supernatant , leaving 100 μl of the supernatant to aspirate the suspended bacterial pellet with a gun, and then evenly spread it on an LB plate containing 100 mg / L spectinomycin. Colonies were visible in about 2 days. Pick a single colony, inoculate it in 5 ml of LB liquid medium containing 100 mg / L spectinomycin, an...
Embodiment 3
[0070] Example 3. Transient Infestation of Tobacco
[0071] 1. Agrobacterium preparation: Take the Agrobacterium preserved at -80°C in Example 2, streak it on an LB plate containing 100 mg / L spectinomycin, and culture it at 28°C for 2 days. Pick a single colony and inoculate it in 5ml LB liquid medium containing the same concentration of antibiotics, and shake at 200rpm at 28°C for 16h. Take 5ml of the bacterial solution and add it to a new 50ml LB medium, shake and culture at 200rpm 28°C for about 6-8h to make the OD600 value 0.6-0.8. Centrifuge at 5000rpm for 15min at 4°C, and use 50ml of resuspension (MS+ 100 μM AS) at 180 rpm at 28° C. for 1 hour before use.
[0072] 2. Agrobacterium infection and co-cultivation: Take the healthy leaves of the aseptic tobacco seedlings and cut them into about 0.5cm 2 size, immersed in the ready-to-use Agrobacterium suspension for 20 minutes. After the end, the excess suspension was sucked off with filter paper, transferred to MS solid m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com