Patents
Literature
74 results about "Population screening" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Population screening refers to a test that is offered to all individuals in a target group, usually defined by age, as part of an organised program.
Apparatus and method for noninvasive monitoring of analytes in body fluids
InactiveUS7214190B1ModificationMaterial analysis by electric/magnetic meansIntravenous devicesMedication infusionNMR - Nuclear magnetic resonance
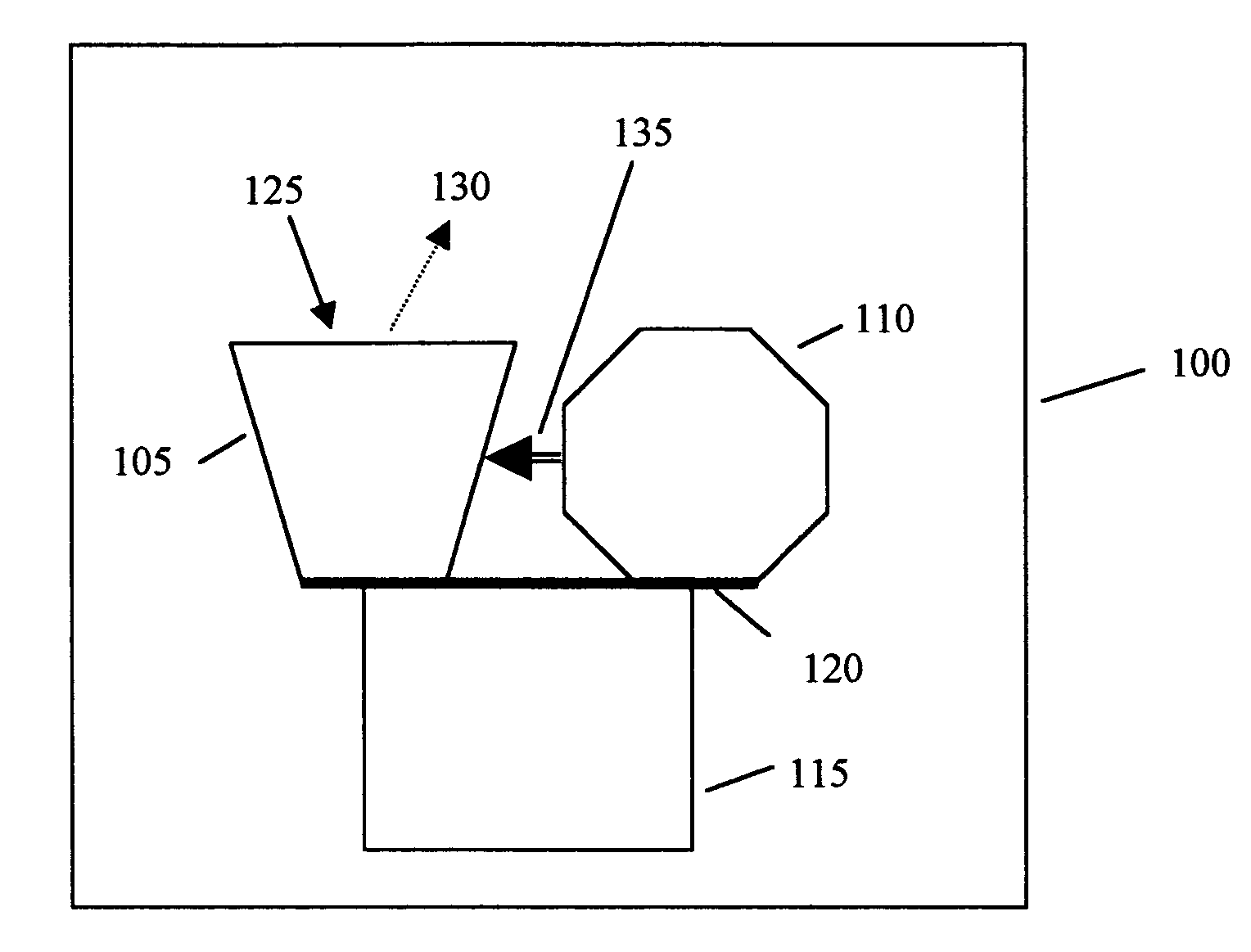
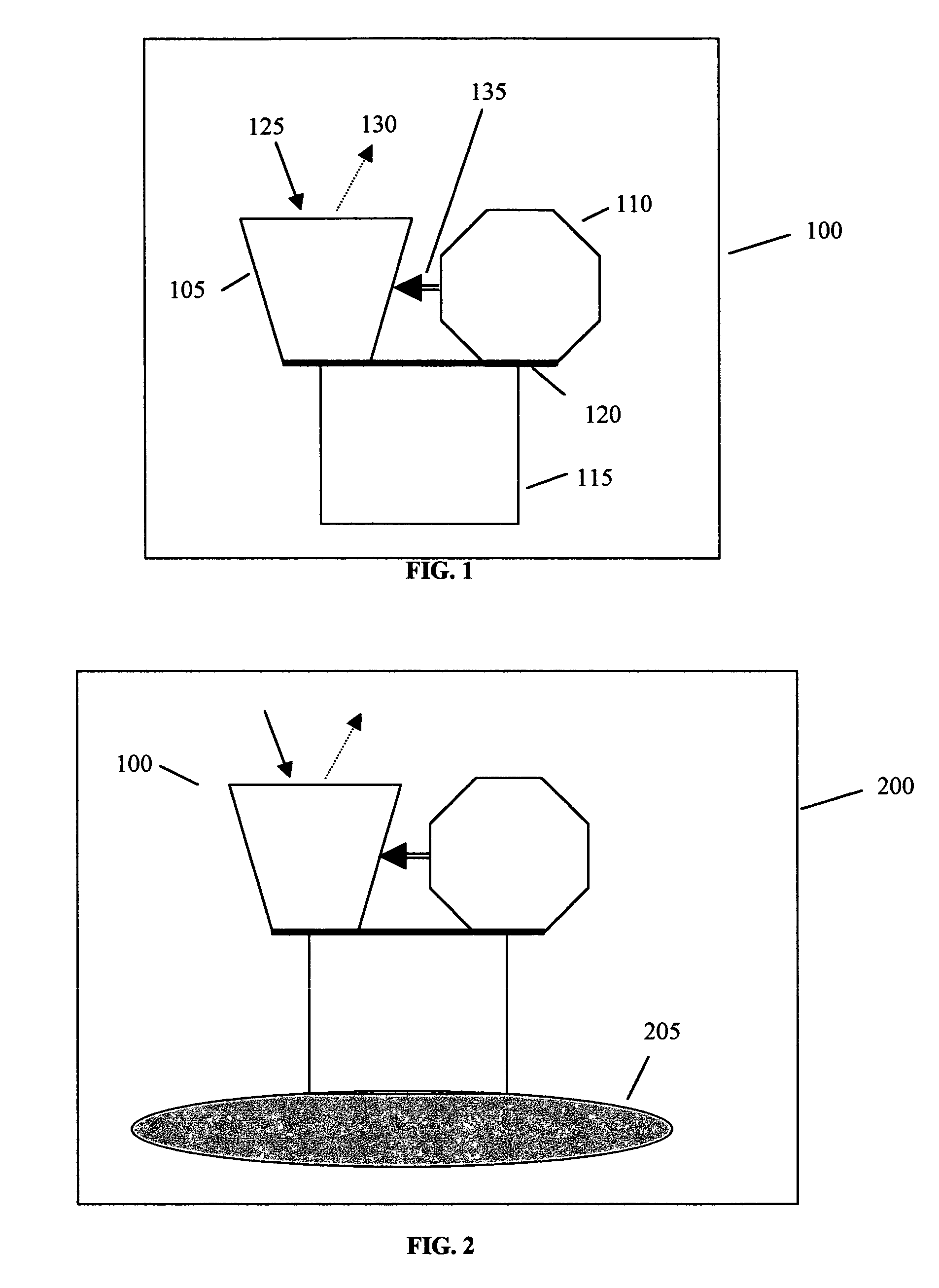
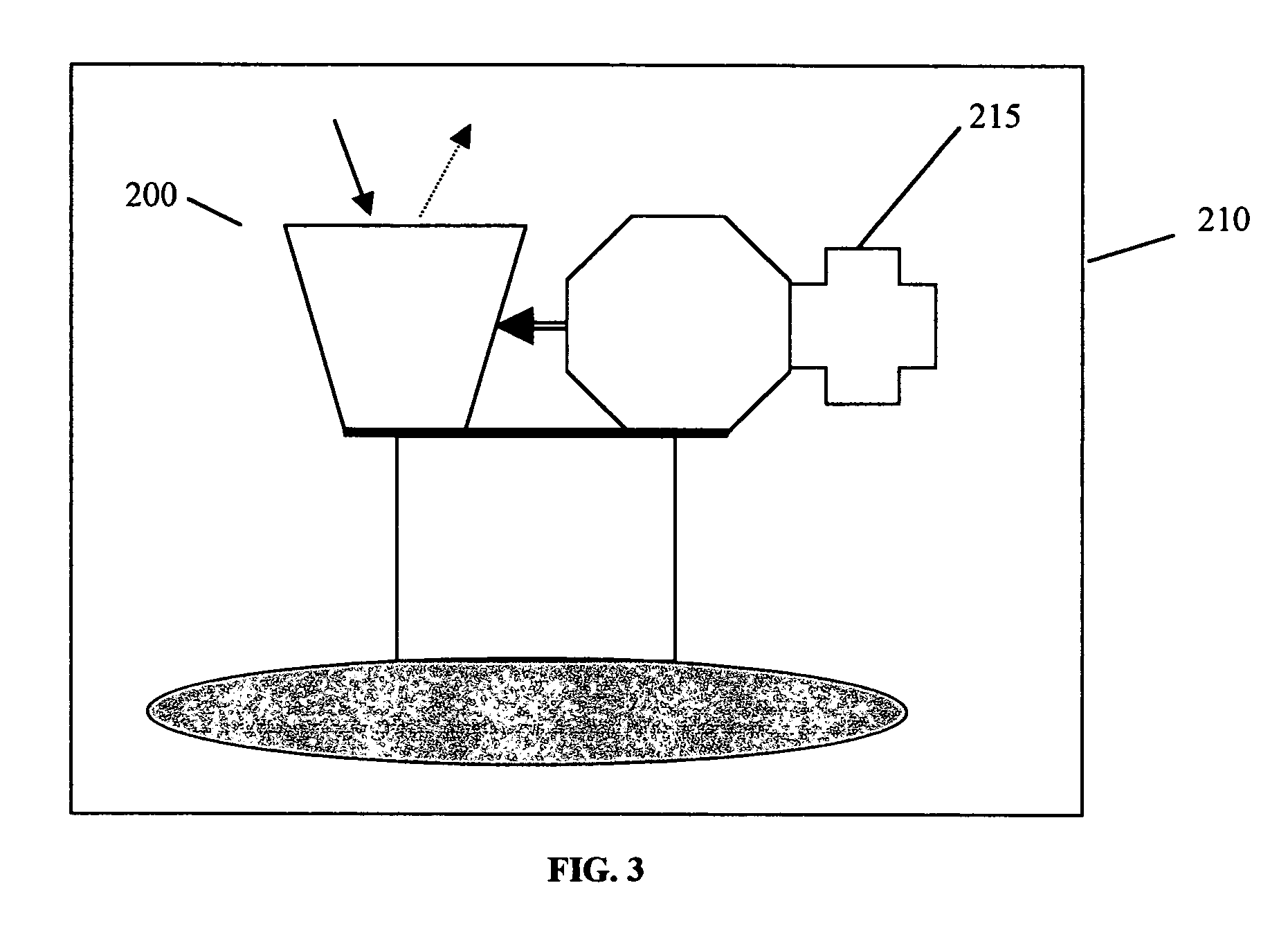
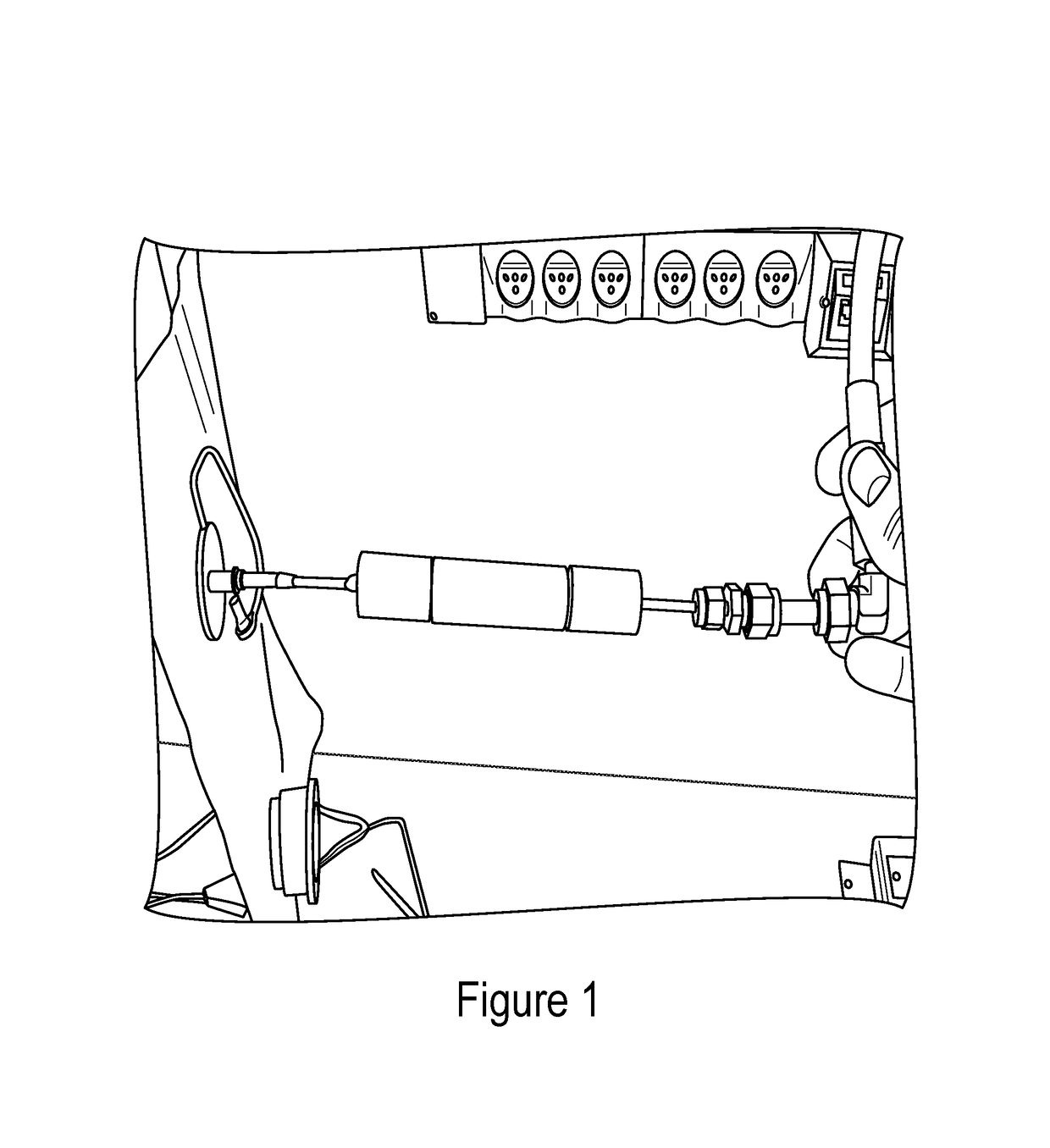
Noninvasive in vivo real time analyte measurement uses a multitude of sensors binding reversibly to the analyte whereby the response of the sensors to a noninvasive stimulus is altered by their bound versus unbound state. The stimulus and responses are electromagnetic, magnetic or any other suitable forms. The sensors are bound to a blood component providing transport through the body fluids and sensor elimination. A sensor is constructed from proteins or as a nanodevice. A noninvasive device generates the stimulus, senses the responses, determines the measurement, and controls a medication infusion pump. A non-contact device is used for population screening, and one form of such a device is a nuclear magnetic resonance imager. Measurement in fluids other than blood uses a blood component flowing out of blood and into the desired fluid.
Owner:WILSON KITCHENER CLARK
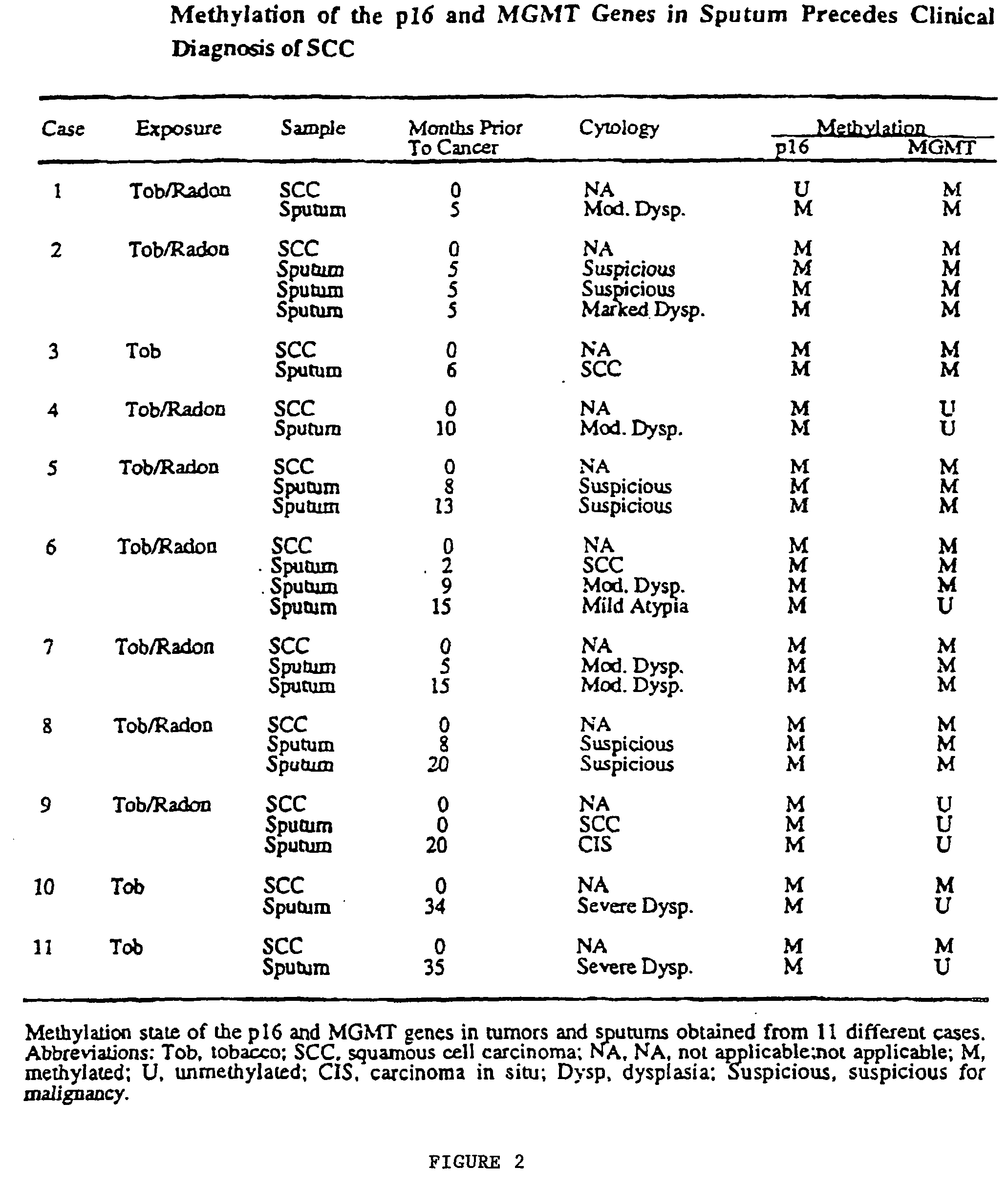
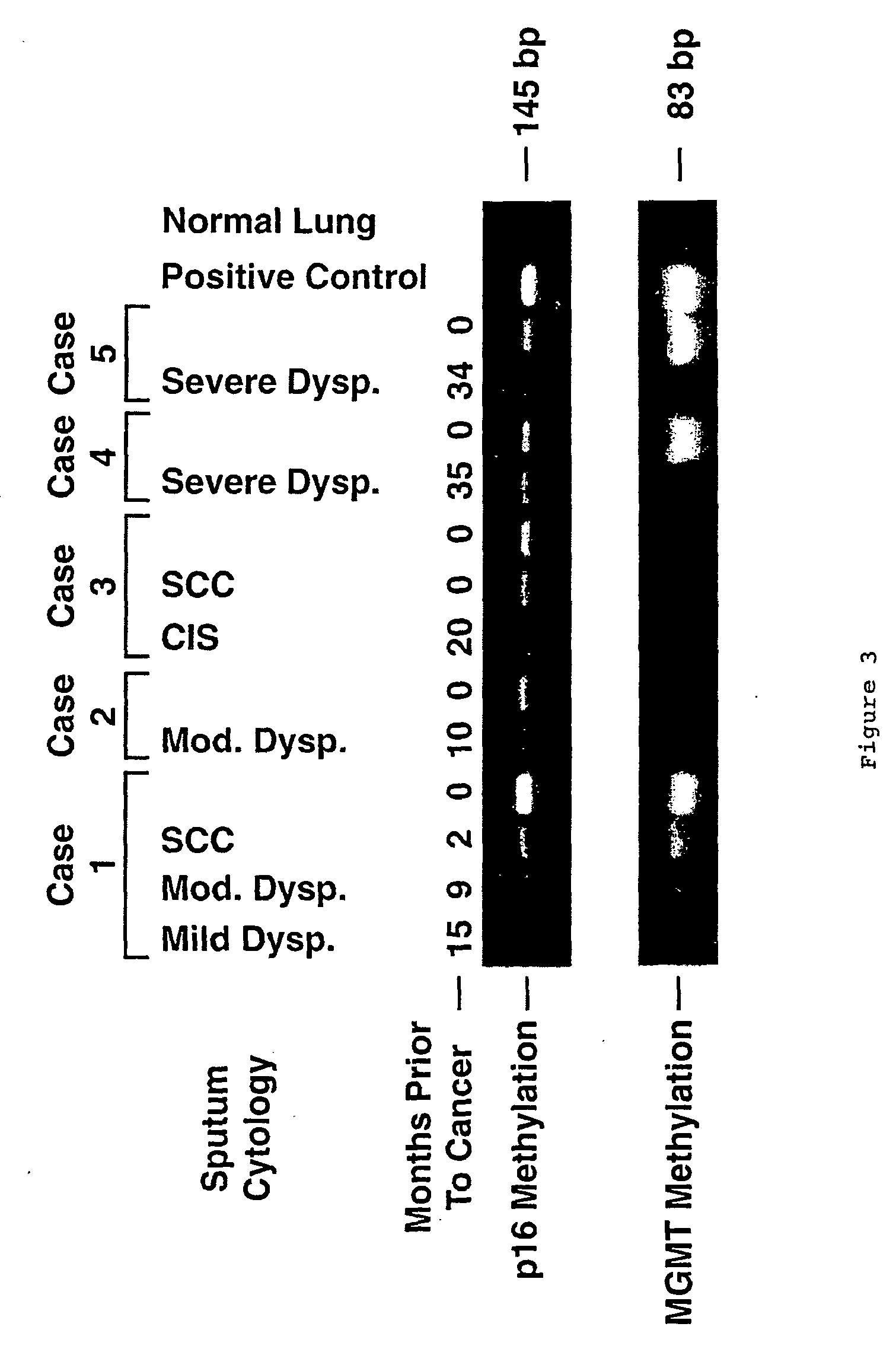
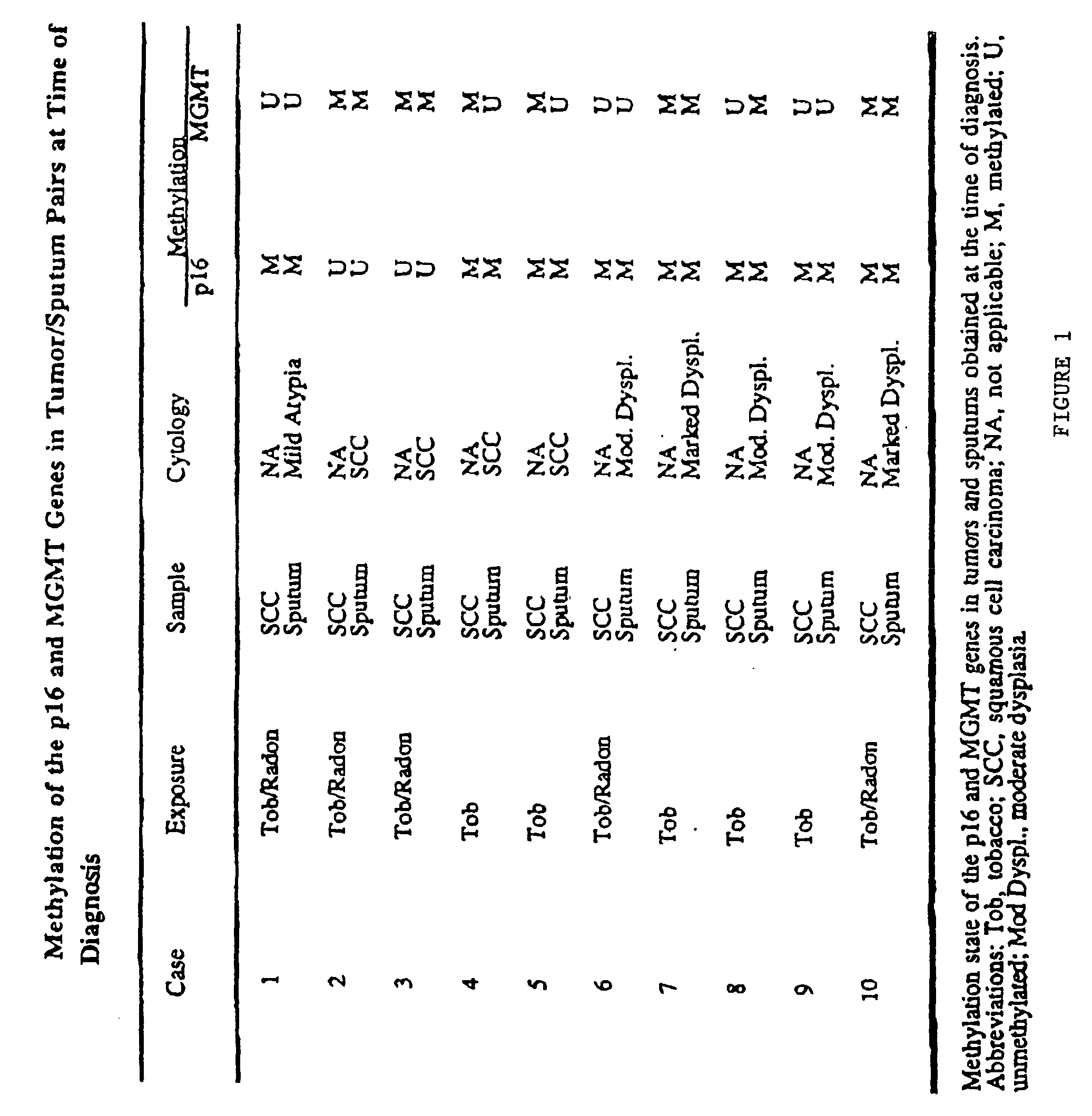
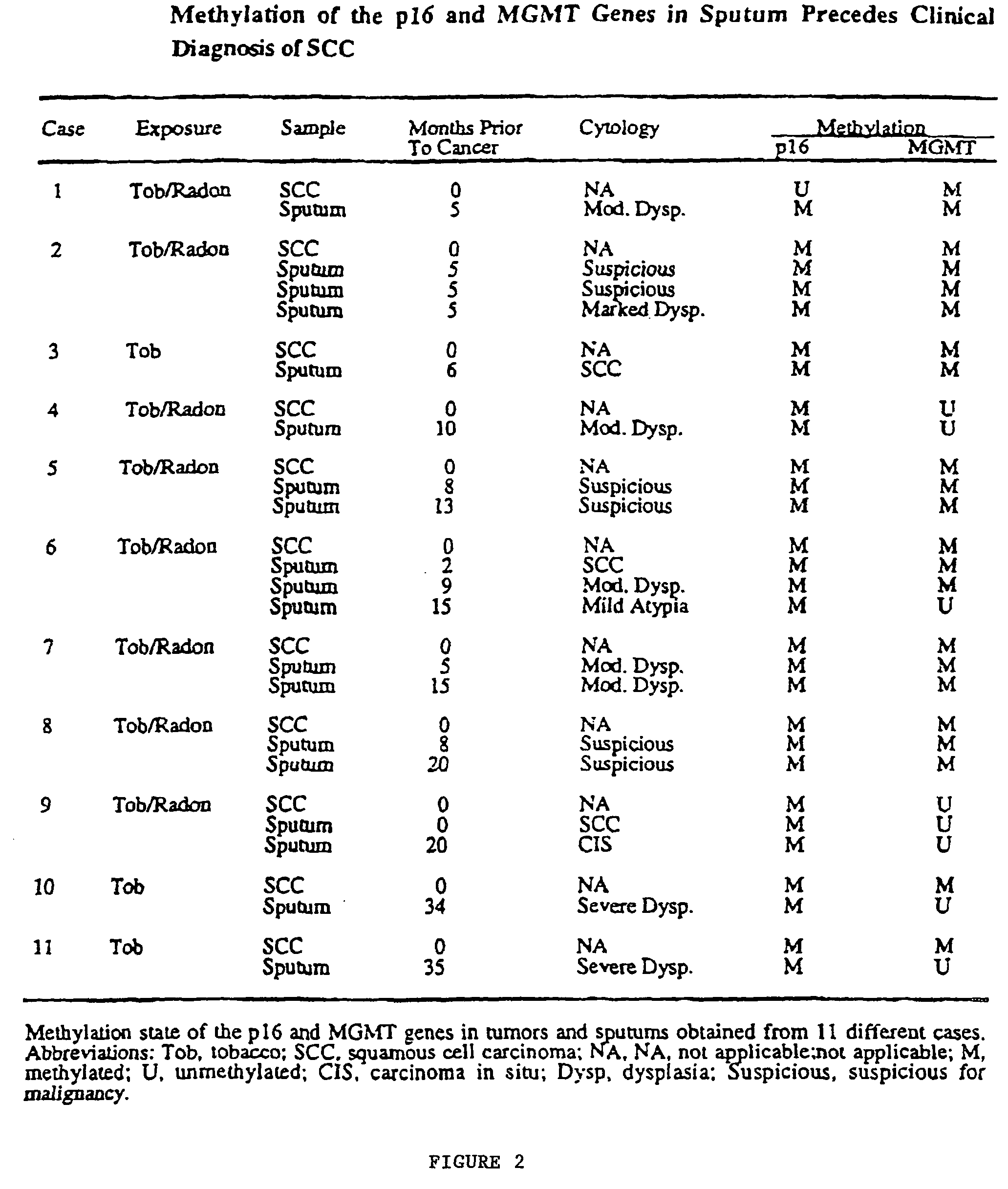
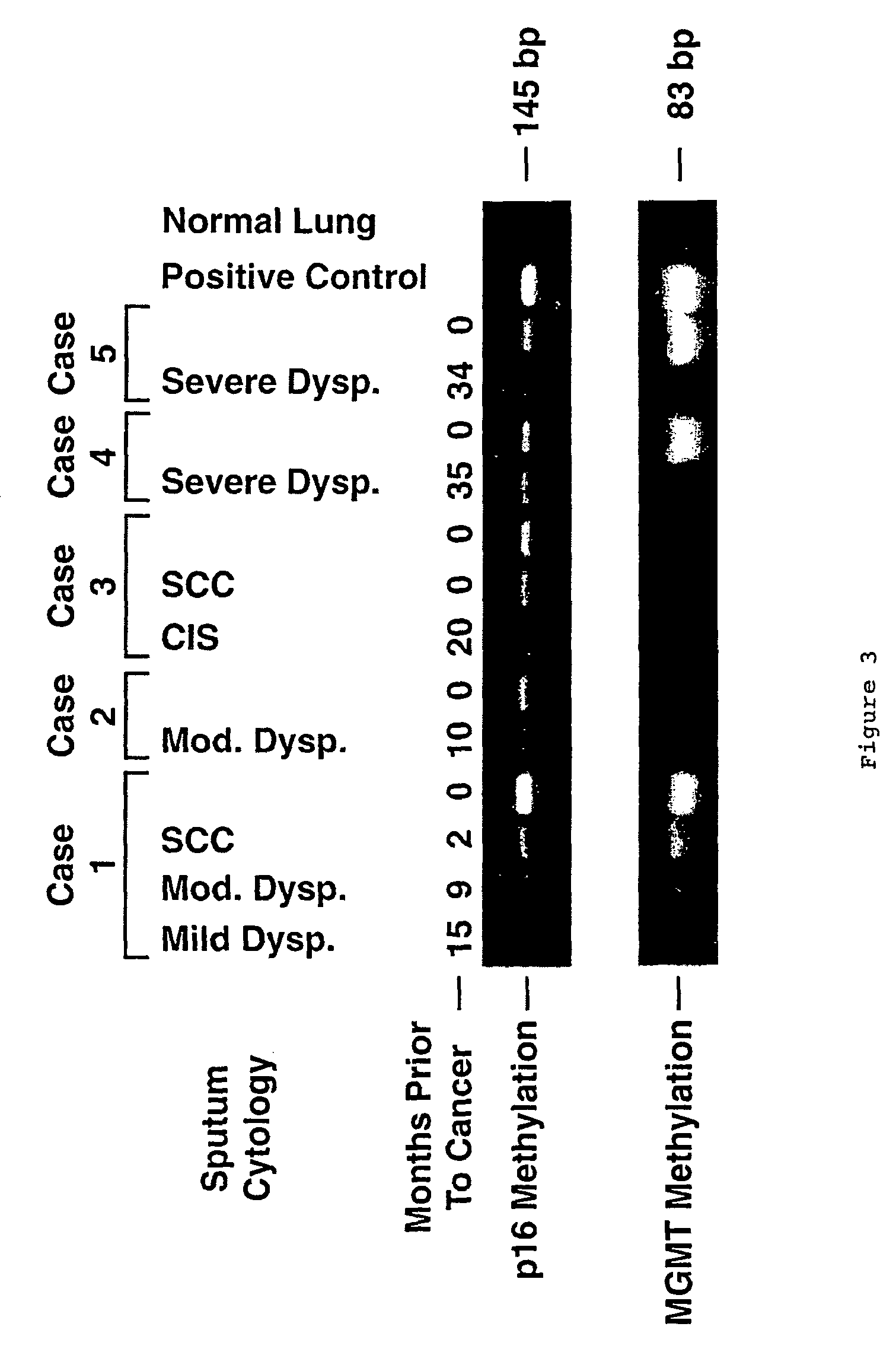
Nested methylation-specific polymerase chain reaction cancer detection method
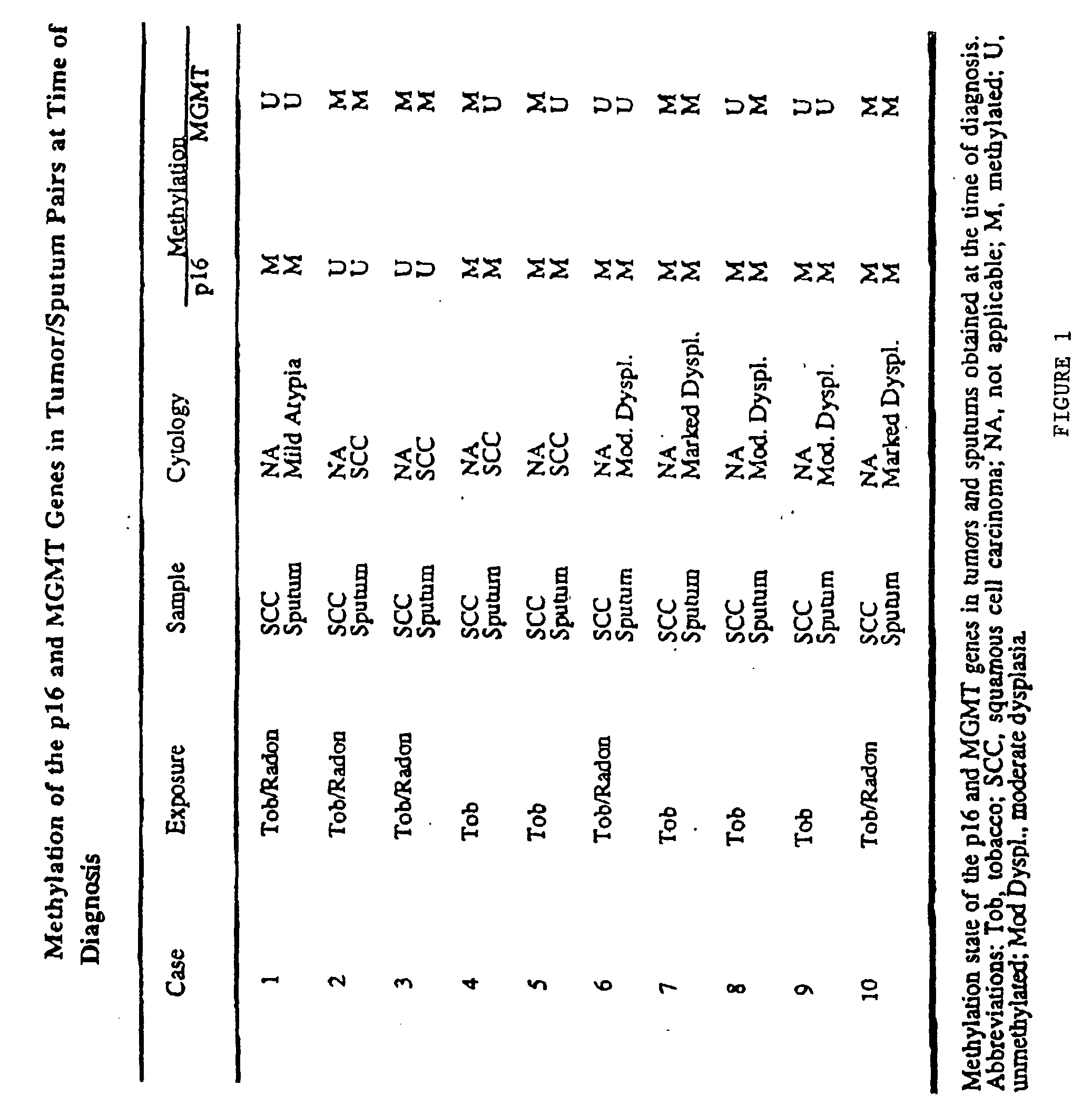
A molecular marker-based method for monitoring and detecting cancer in humans. Aberrant methylation of gene promoters is a marker for cancer risk in humans. A two-stage, or "nested" polymerase chain reaction method is disclosed for detecting methylated DNA sequences at sufficiently high levels of sensitivity to permit cancer screening in biological fluid samples, such as sputum, obtained non-invasively. The method is for detecting the aberrant methylation of the p16 gene, O 6-methylguanine-DNA methyltransferase gene, Death-associated protein kinase gene, RAS-associated family 1 gene, or other gene promoters. The method offers a potentially powerful approach to population-based screening for the detection of lung and other cancers.
Owner:LOVELACE RESPIRATORY RES INST
Preparation method of non-transgenic CRISPR mutant
InactiveCN108611364AReduce frequencyImprove accuracyNucleic acid vectorVector-based foreign material introductionScreening methodSexual reproduction
The invention provides a preparation method of a non-transgenic mutant based on a CRISPR-Cas9 technology. Specifically, the preparation method comprises a construction method and a screening method. The construction method is characterized in that CRISPR-Cas9 and sgRNA are preassembled and are located at T-DNA of agrobacterium tumefaciens; site-directed change of a target gene of a target plant can be realized by infection of the target plant by the agrobacterium tumefaciens and coculture without integrating the sequences of the CRISPR-Cas9 and the sgRNA into a genome of the target plant, so that an obtained site-directed mutant material of the target gene does not need the processes of sexual reproduction, segregation posterity, population screening and the like or does not contain any exogenous gene sequence. The obtained regeneration seedlings are screened by a high-throughput sequencing and high-resolution melting curve technology provided by the invention; mutant plants of which target genes are mutated can be obtained by identifying the regeneration seedlings efficiently and quickly at the current generation of transgene, even if low-proportion mutants of which the proportionis 1 / 100 of a mixed population can be screened. The screening method provided by the invention still can ensure excellent accuracy and sensitivity.
Owner:NANJING AGRICULTURAL UNIVERSITY
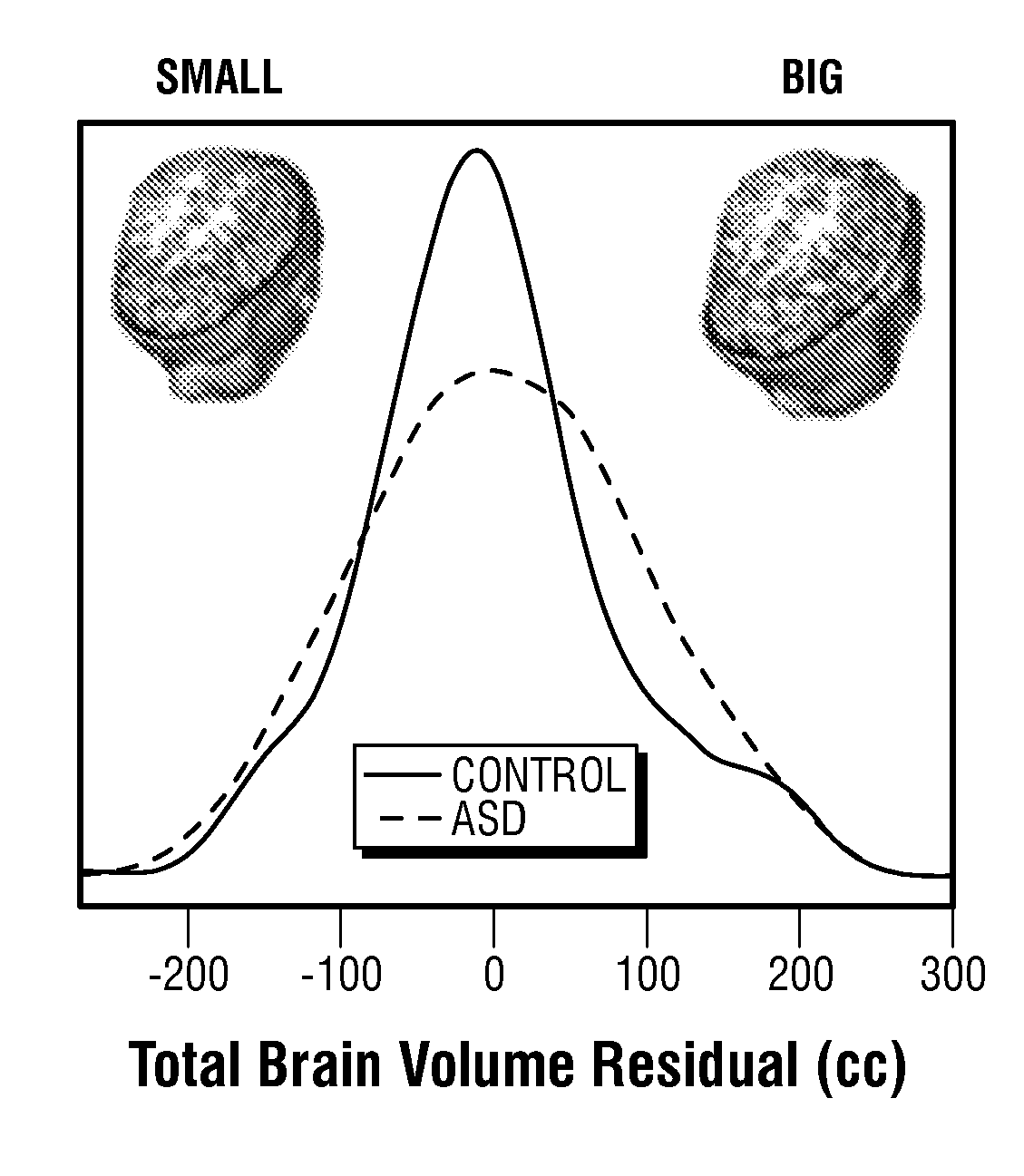
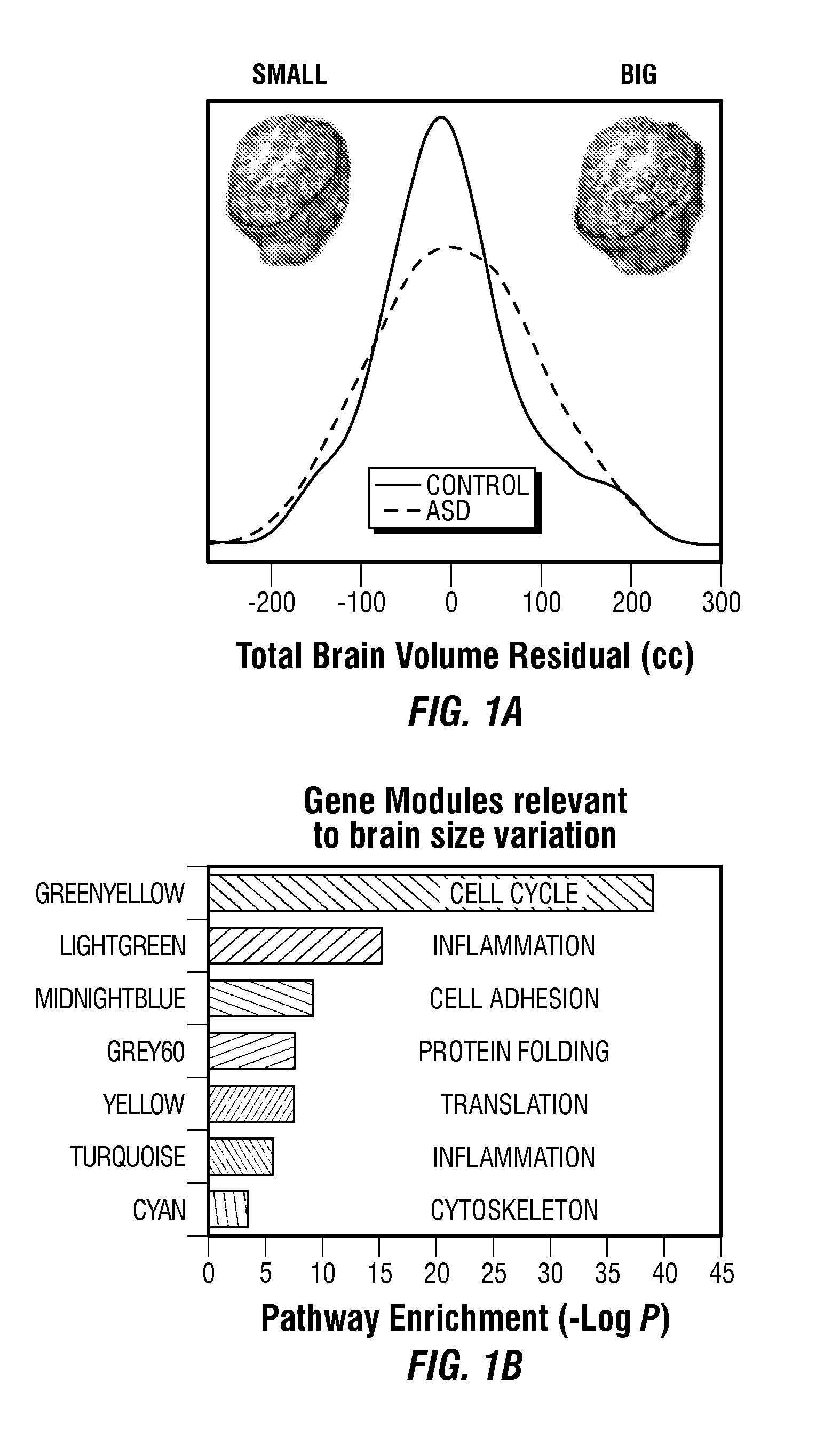
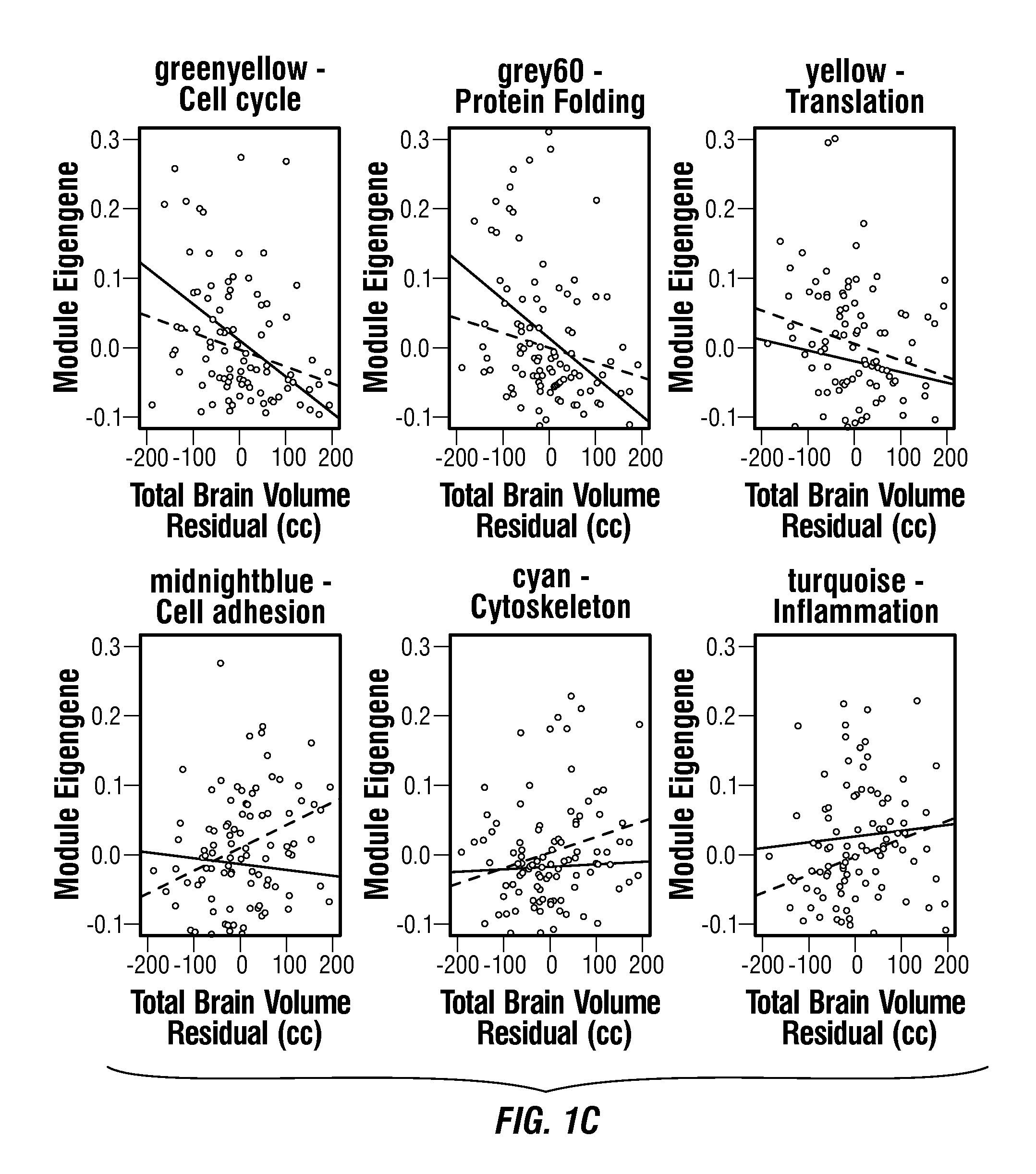
Screening, Diagnosis and Prognosis of Autism and Other Developmental Disorders
ActiveUS20150227681A1Improve accuracyHigh sensitivityMicrobiological testing/measurementLibrary screeningDevelopmental disorderCrowds
The invention provides a method and system combining functional genomic and genetic, proteomic, anatomic neuroimaging, functional neuroimaging, behavioral and clinical measurements and data analyses for autism pediatric population screening, diagnosis or prognosis. More specifically, the invention provides a weighted gene and feature test for autism which uses a weighted gene signature matrix for comparison to a reference database of healthy and afflicted individuals. The invention also provides normalized gene expression value signatures for comparison to a reference database. The invention additionally combines either the weighted gene or the normalized gene analysis with comparisons to a gene-networks signature matrix, a multi-modal signature matrix, and a collateral features signature matrix for improved accuracy in screening, diagnostic and prognostic relevance for autism, particularly for newborns, babies ages birth to 1 year, toddlers ages 1 to 2 years, toddlers ages 2 to 3 years and young children ages 3 through 4 years.
Owner:RGT UNIV OF CALIFORNIA
Method for detecting gene copy number variation
ActiveCN102409088ASimple designThe result is accurateMicrobiological testing/measurementPrenatal diagnosisWild type
Owner:郭奇伟 +1
Method and kit for diagnosing bladder cancer with urine
InactiveCN102311953AMicrobiological testing/measurementDNA/RNA fragmentationCommunity health centerBiomarker (petroleum)
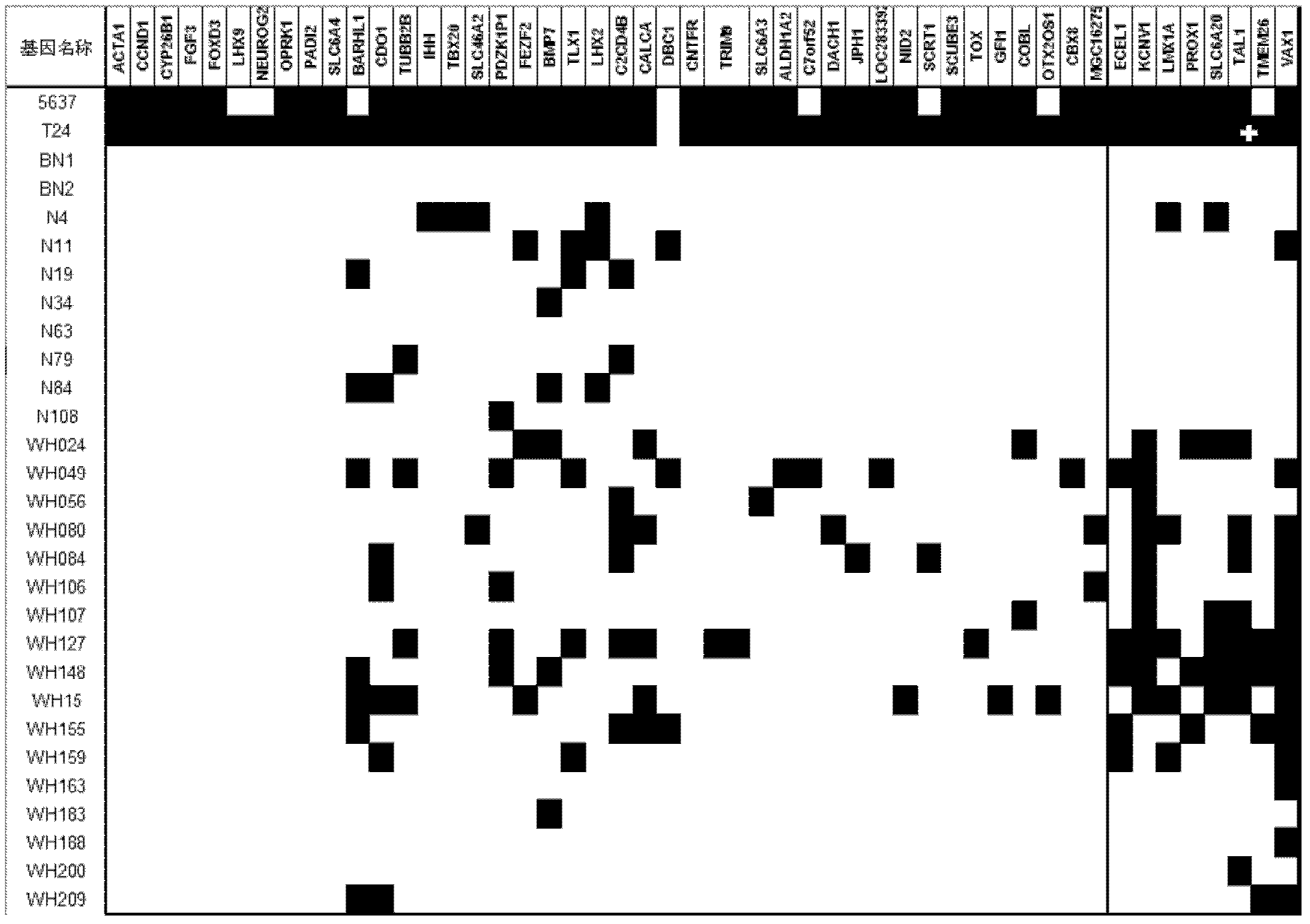
The invention relates to a method and a kit for diagnosing bladder cancer with urine. The invention discloses a group of methylated sensitive genes, comprising ECEL1, KCNV1, LMX1A, PROX1, SLC6A20, TAL1, TMEM 26, and VAXI gene. In urine samples of bladder cancer patients, the specific CpG sites of the genes are showed the highest methylation levels. Accordingly, the genes are the biological marker of bladder cancer. The method and the kit can be used as the basic of designing diagnostic reagents of bladder cancer, and are suitable for auxiliary detection of cancer in hospitals, postoperative followup, screening of high risk group of bladder cancer in community health centers, and screening of common people and high risk practitioners of bladder cancer in physical examination center.
Owner:SHANGHAI INST OF ONCOLOGY
Sensor Technology for Diagnosing Tuberculosis
ActiveUS20150301021A1Enhanced sensitivity and selectivityFast and reliable diagnosisVibration measurement in solidsImmobilised enzymesMedicineNanoparticle
A sensor technology comprising a single nano-material (gold nanoparticles and / or carbon nanotube) based sensor or a plurality of sensors in conjunction with a pattern recognition algorithm for non-invasive and accurate diagnosis of tuberculosis caused by M. tuberculosis bacteria in a subject. The sensor technology is suitable for population screening of tuberculosis, particularly in resource-poor and developing countries.
Owner:TECHNION RES & DEV FOUND LTD
Autoantibody joint detection ELISA kit for screening early esophageal cancer
ActiveCN110187108AEfficient detectionHigh detection sensitivityMaterial analysisHigh risk populationsElisa kit
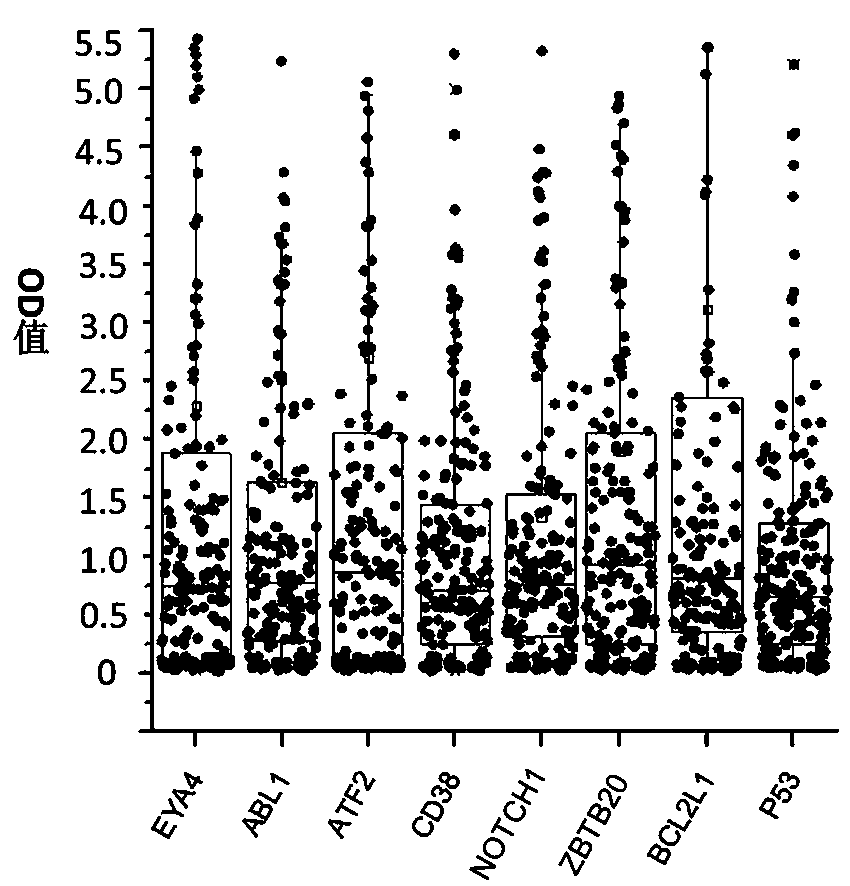
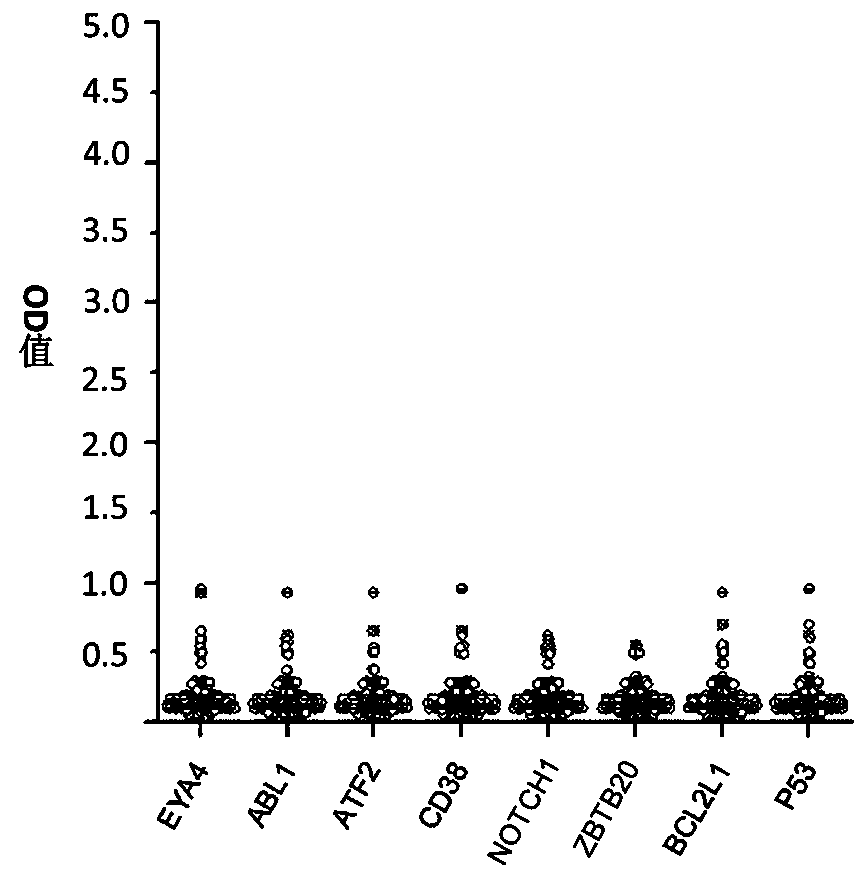
The invention belongs to the technical field of tumor medicine, and particularly discloses an autoantibody joint detection ELISA kit for screening early esophageal cancer. The kit comprises a solid-phase carrier and tumor-associated antigens coated on the solid-phase carrier, wherein the tumor-associated antigens are EYA4, ABL1, ATF2, CD38, NOTCH1, ZBTB20, BCL2L1 and P53. Furthermore, the kit alsocomprises a sample diluent, a second antibody, a second antibody diluent, positive control serum, negative control serum, a color developing solution, a stop solution and a washing solution. The ELISA kit provided by the invention can effectively detect esophageal cancer, especially early esophageal cancer, has the detection sensitivity of 94% and the specificity of 79%, can be used for large-scale screening of asymptomatic people in high-incidence areas of esophageal cancer, and is beneficial to screening and early discovery of asymptomatic high-risk population.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV +1
Nested methylation-specific polymerase chain reaction cancer detection method
A molecular marker-based method for monitoring and detecting cancer in humans. Aberrant methylation of gene promoters is a marker for cancer risk in humans. A two-stage, or “nested” polymerase chain reaction method is disclosed for detecting methylated DNA sequences at sufficiently high levels of sensitivity to permit cancer screening in biological fluid samples, such as sputum, obtained non-invasively. The method is for detecting the aberrant methylation of the p16 gene, O 6-methylguanine-DNA methyltransferase gene, Death-associated protein kinase gene, RAS-associated family 1 gene, or other gene promoters. The method offers a potentially powerful approach to population-based screening for the detection of lung and other cancers.
Owner:LOVELACE RESPIRATORY RES INST
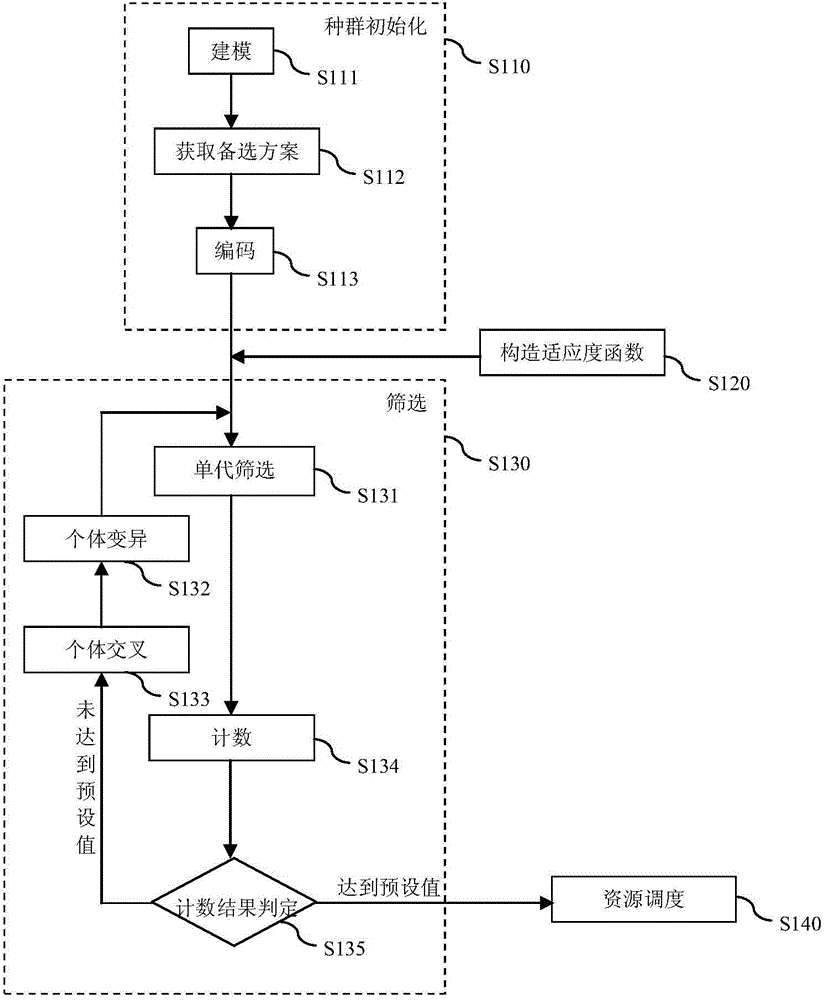
Resource scheduling method for cloud system
ActiveCN104899100AGood load balancing resultsSmall migration costResource allocationPopulation screeningDistributed computing
The present invention discloses a resource scheduling method for a cloud system. The method comprises the following steps of: in a population initialization step, an alternative scheme of resource scheduling of the cloud system is obtained, and initial population is created by utilizing the alternative scheme; in a fitness function creation step, a total fitness function is created for specific demands of resource scheduling of the cloud system; in a population screening step, individuals in the initial population are screened by utilizing the total fitness function to obtain fitted individuals; and in a resource scheduling step, a virtual machine and a node controller are subjected to resource scheduling according to the alternative scheme corresponding to the fitted individuals. Compared to the prior art, the resource scheduling method disclosed by the present invention is relatively low in transfer cost while realizing good load balancing result.
Owner:BEIJING UNIV OF POSTS & TELECOMM
Joint detection kit for alpha,beta-thalassemia associated mutant genes
ActiveCN102181560AHigh detection specificityLow costMicrobiological testing/measurementPrenatal diagnosisBeta thalassemia
The invention discloses a joint detection kit for detecting alpha,beta-thalassemia associated with mutant genes, which comprises (1) a gene chip and (2) a primer, wherein the gene chip is provided with probes, and the probes are a sequence SEQ ID Nos:1-31 and a sequence which is complementary to the sequence SEQ ID Nos:1-31; and the primer is a sequence SEQ ID Nos:32-42. The thalassemia gene detection kit provides a platform for detecting 16 mutant genes associated with alpha-thalassemia (three deletion types and two mutant types) and beta-thalassemia, can perform synchronous joint detection,improve the specificity of detection, reduce cost and shorten detection time, and has a great significance for the screening of patients suffering from thalassemia, genetic counseling and prenatal diagnosis.
Owner:潮州凯普生物化学有限公司 +1
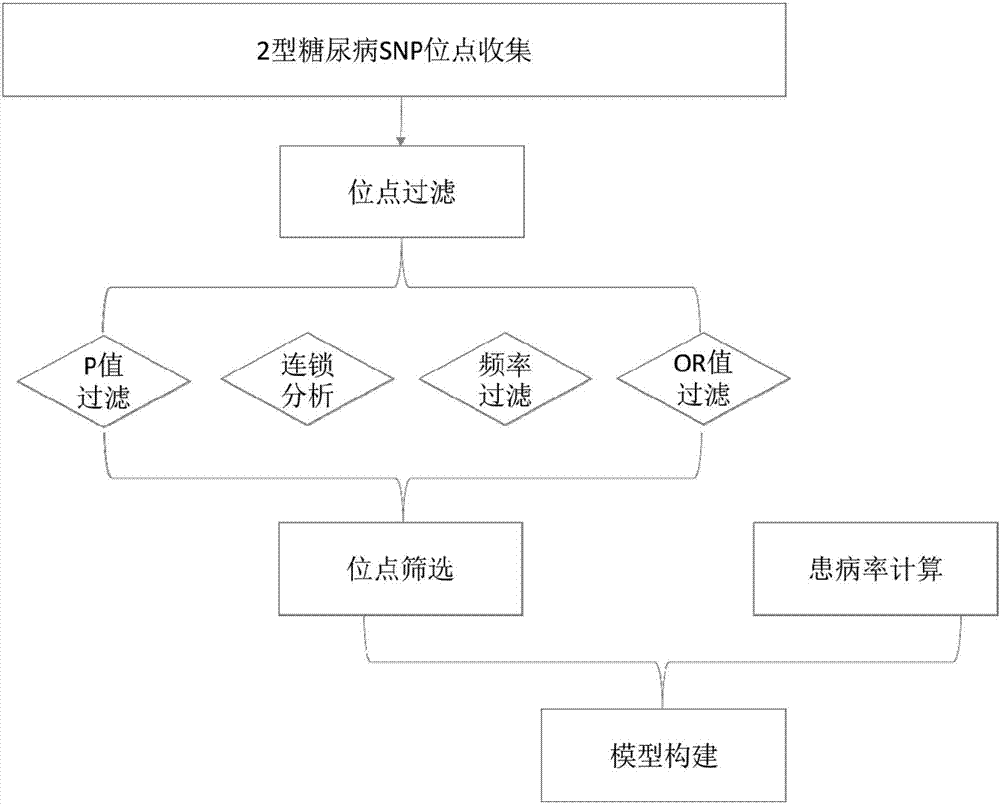
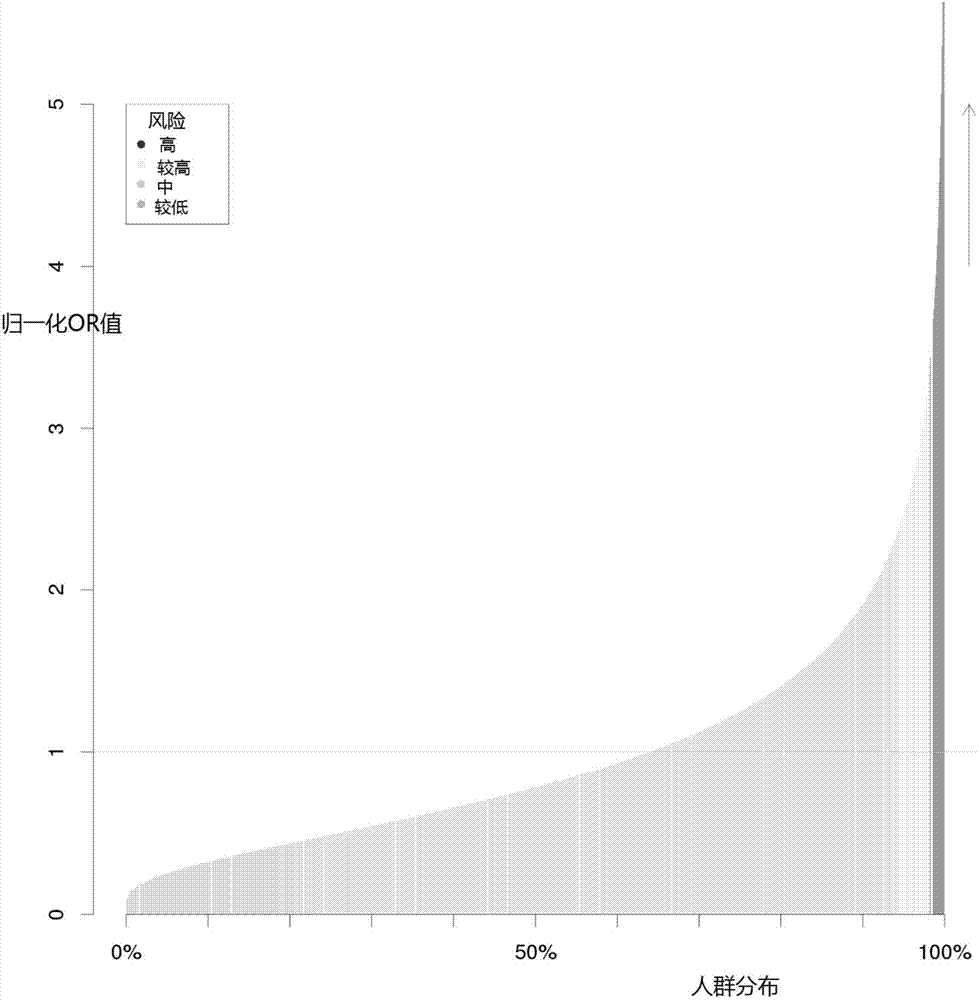
Construction method and construction system for type 2 diabetes mellitus risk assessment model
InactiveCN107256323AAccurate calculationReduce morbiditySpecial data processing applicationsHigh risk populationsNucleotide
The invention discloses a construction method and a construction system for a type 2 diabetes mellitus risk assessment model. The method comprises the following steps that: (1) selecting an SNP (Single Nucleotide Polymorphisms) locus related to type 2 diabetes mellitus; (2) calculating the risk degree, i.e., an OR (Odds Ratio) value, of the SNP locus; (3) calculating the frequency of the SNP locus in an East Asia population; (4) calculating the epidemiology prevalence rate of the type 2 diabetes mellitus; and (5) according to a Bayesian algorithm and a Hardy-Weinberg equilibrium principle, constructing the risk assessment model. The model of the invention calculates the mathematical expectation value and the OR value of the risk allele amount of the population, the epidemiology prevalence rate of the type 2 diabetes mellitus is combined to obtain the average prevalence rate and the confidence interval of a group on the basis of inheritance, a new construction method for the type 2 diabetes mellitus risk assessment model is provided, so that the prevalence risk of the type 2 diabetes mellitus more approaches to a true situation, and a result is more scientific and reasonable. By use of the method, high-risk population screening accuracy is improved, the prevalence rate of the type 2 diabetes mellitus is expected to be lowered, and a great quantity of expenditures can be saved for the nation and society so as to benefit the nation and the people.
Owner:云健康基因科技(上海)有限公司
Method for constructing serum metabonomics analysis model
ActiveCN104713970AAchieve the purpose of early screeningA large amountComponent separationSerum samplesPrincipal component analysis
The invention discloses a method for constructing a serum metabonomics analysis model. The construction method comprises the following steps: collecting a healthy serum sample and an ill serum sample; performing LC-MS detection on the samples, thereby obtaining original metabolic fingerprints; preprocessing the fingerprints, sequentially performing principal component analysis and partial least square discriminant analysis on a two-dimensional matrix, thereby obtaining a PLS-DA model; and verifying the obtained PLS-DA model, wherein if the overfitting risk does not exist, the model construction is finished. According to the method disclosed by the invention, the serum metabonomics analysis technology is applied to early screening of an esophagus cancer, the high-risk population of the esophagus cancer can be rapidly and conveniently screened by virtue of model construction, the esophagus cancer screening range is widened, the entire population screening efficiency in the high incidence area of esophagus cancer is effectively improved, the screening cost is greatly reduced, pain of partial population caused by an invasive gastroscope is effectively avoided, and the method has significant economic and social benefits and is worthy of popularization and application.
Owner:SHANDONG TUMOR HOSPITAL
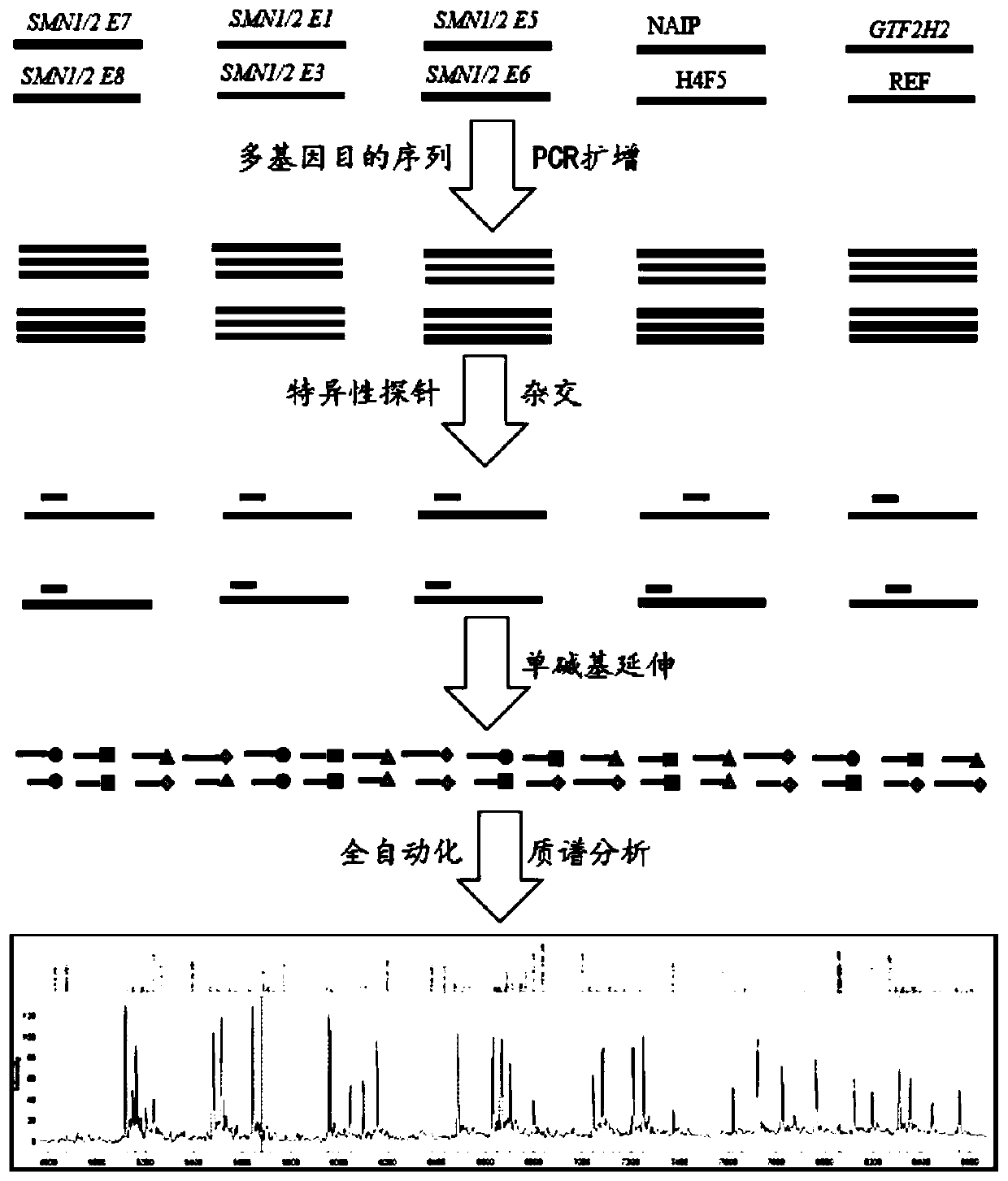
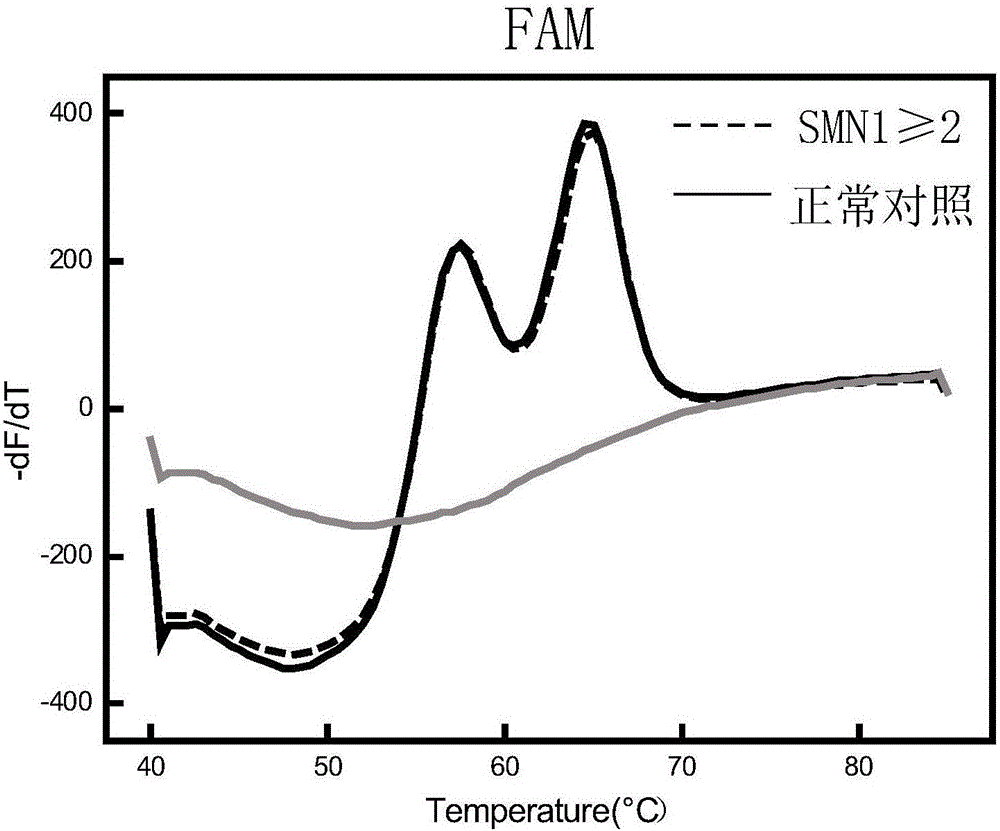
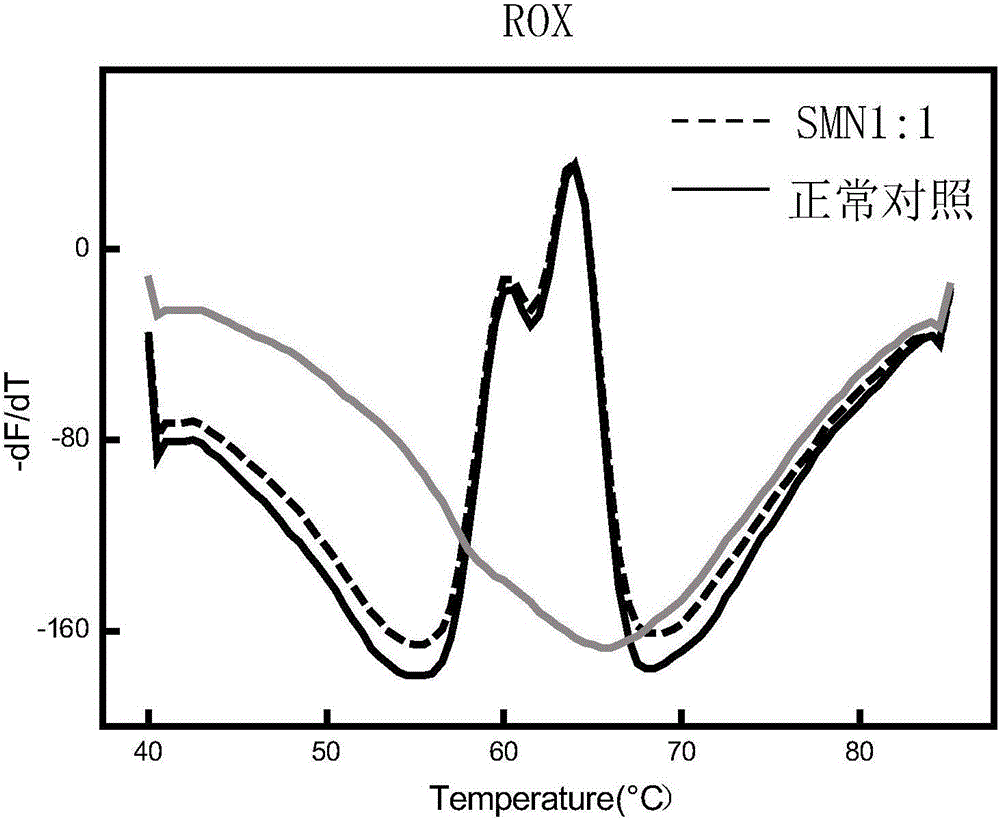
Time-of-flight mass spectrometry nucleic acid analysis method for detecting human spinal muscular atrophy gene mutation
ActiveCN110468192AImprove stabilityImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationPrenatal diagnosisTime of flight
The invention discloses a time-of-flight mass spectrometry nucleic acid analysis method for detecting human spinal muscular atrophy gene mutation. A primer combination comprising an amplification primer and a mass spectrometry extension probe primer is utilized. According to the method, the copy numbers of sequences related to SMN1, SMN2, NAIP, H4F5 and GTF2H2 genes are quantitatively detected, and whether deletion, deletion number and multiple copies exist or not is analyzed, so that the clinical phenotype severity can be directly deduced; the method has good sensitivity, specificity, stability and accuracy, and effectively solves the technical bottleneck of false negative, false positive and the like; the operation is simple, the cost is relatively low, and the result is stable and reliable; the method is high in flux and low in cost, has general representativeness and universality, is easy to realize automatic and large-scale detection, and is suitable for large-scale population screening; genetic typing detection can be carried out on part of common SMN1 upper point mutations; and the requirements of large-scale population screening, prenatal diagnosis and conventional molecular diagnosis in current SMA prevention and treatment are met.
Owner:GUANGZHOU DARUI BIOTECH
Sensor technology for diagnosing tuberculosis
ActiveUS10168315B2Enhanced sensitivity and selectivityFast and reliable diagnosisDisease diagnosisRespiratory organ evaluationMedicineNanoparticle
Owner:TECHNION RES & DEV FOUND LTD
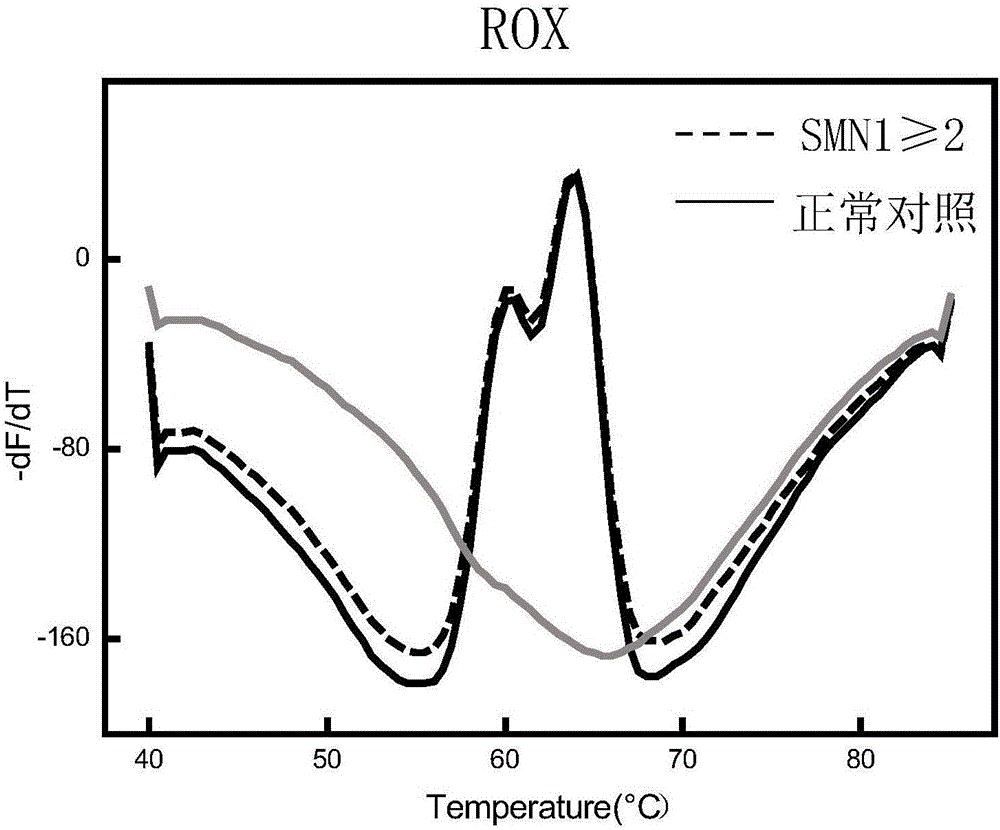
Spinal muscular atrophy pathogenic gene detection kit based on melting curve analysis
The invention relates to a spinal muscular atrophy pathogenic gene detection kit based ona melting curve analysis, and relates to a pathogenic gene detection kit, which comprises an upstream primer F1 and a downstream primer R1 for amplifyingexon7 of SMN1 and SMN2 genes; an upstream primer F2 and a downstream primer R2 for amplifying exon4 of an internal reference CFTR gene; and a fluorescent probe for detection. In a single-tube PCR system, the copy number of exon7 of the SMN1 gene as a main pathogenic gene of SMA can be quantitatively detected, the sample genotype can be known by a fluorescence PCR melting curve analysis after PCR amplification is finished, the whole operation is completed in 2 to 3h, is simple and rapid, and is short in time-consuming; the homogeneous detection and closed tube operation are carried out; the detection flux is high; the detection specificity is high, and the results are easy to interpret. The detection kit can be rapidly and convenientlyapplied to large-scale population screening of the SMA pathogenicgene, and is especially suitable for prenatal, premaritaland pre-pregnancy screening and genetic diagnosis of patients.
Owner:夏众敏 +2
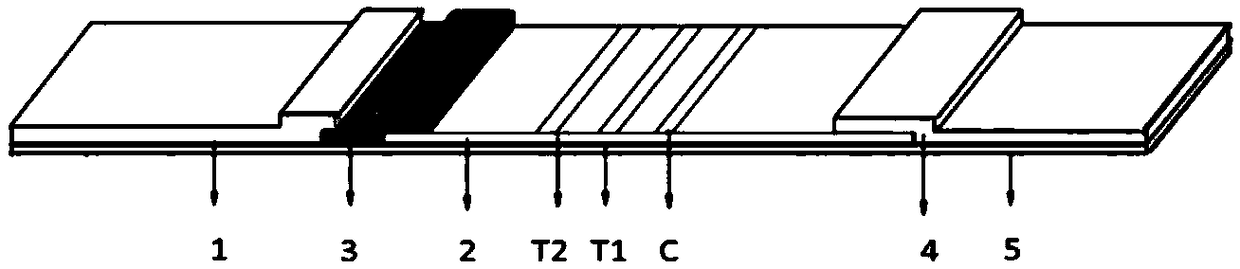
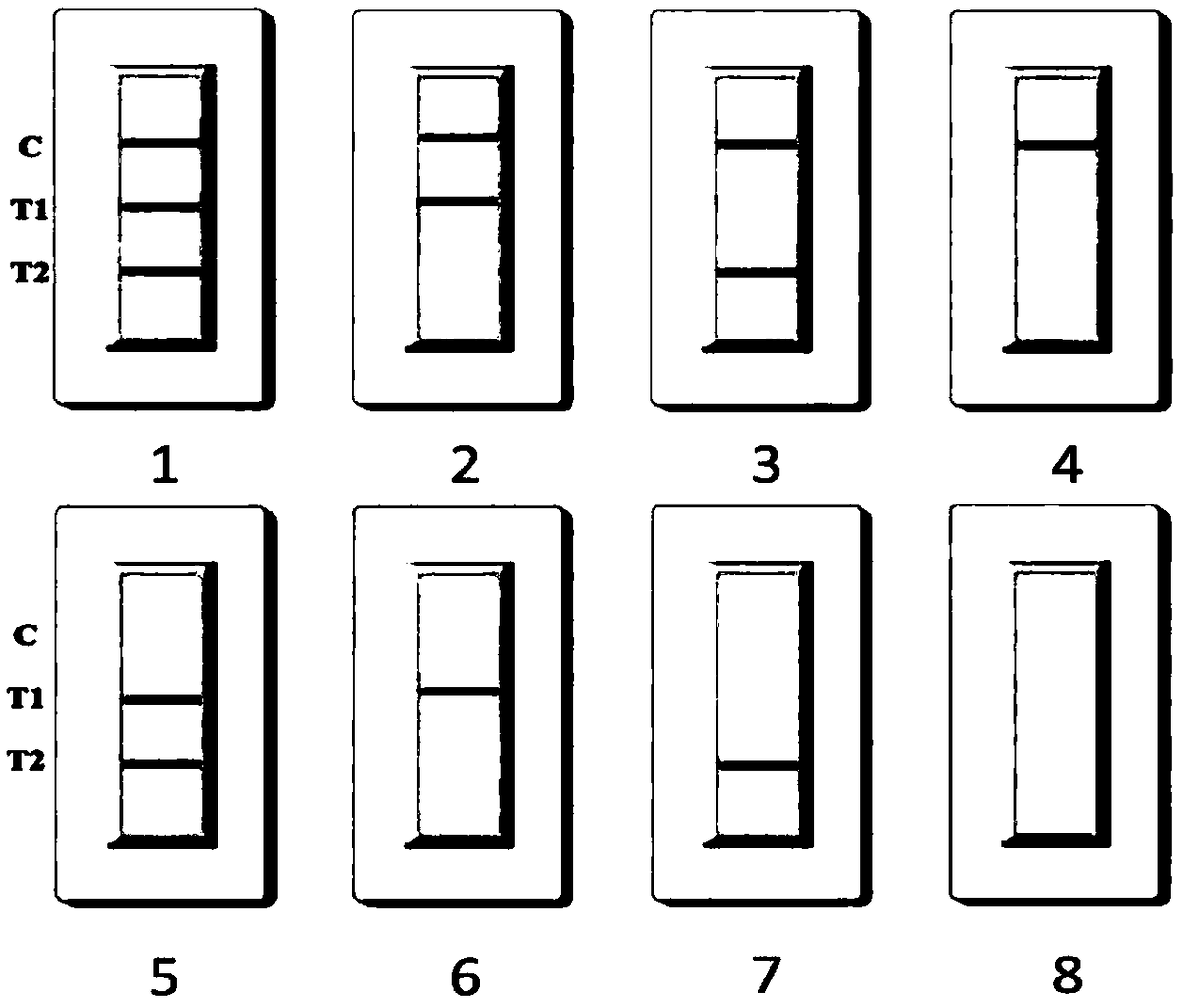
Nano selenium kit for quickly detecting HE4 and CA125
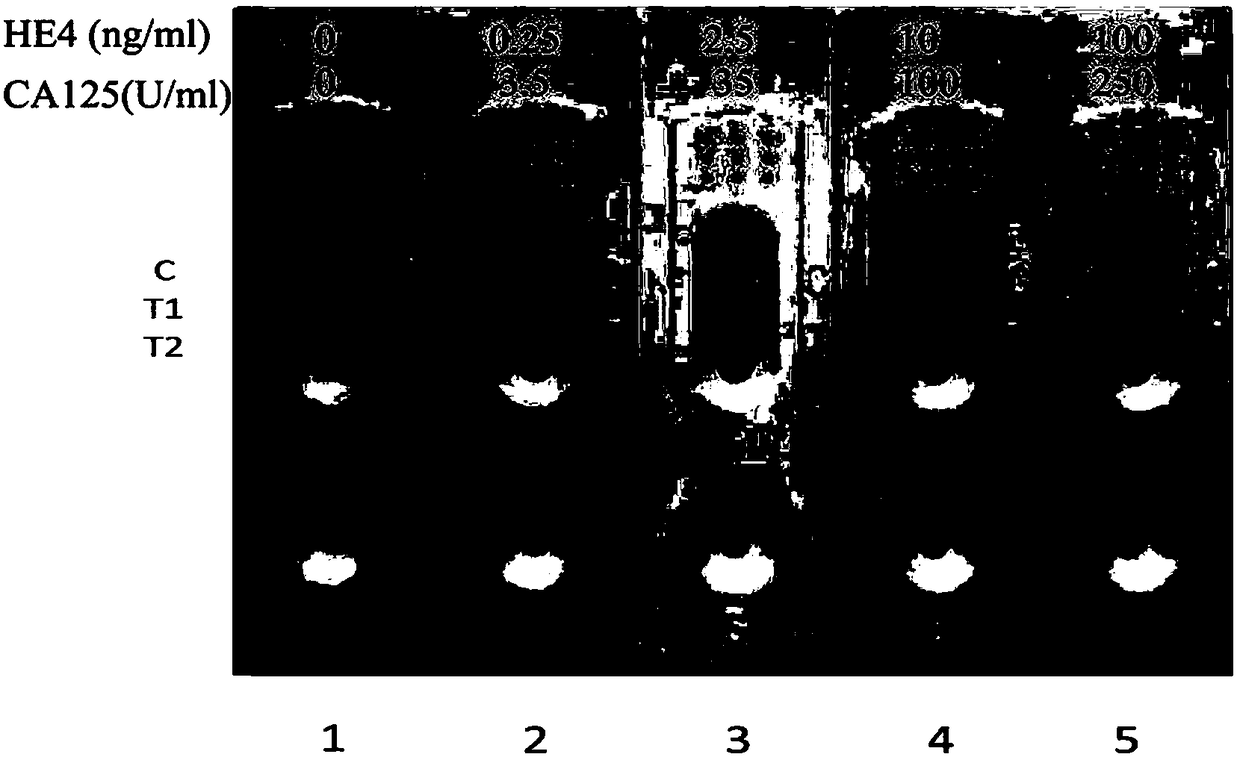
InactiveCN109444435AReduce manufacturing costFast and sensitive screeningDisease diagnosisBiological testingSmall sampleRisk groups
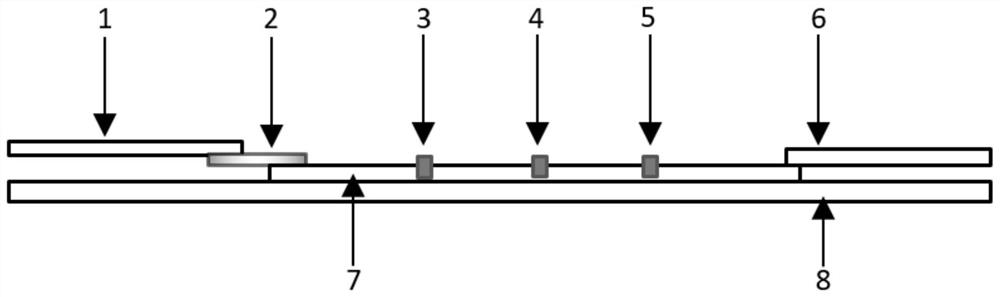
The invention belongs to the field of biological immunochromatography detection methods and specifically relates to a nano selenium kit for quickly detecting HE4 and CA125 and a preparation method thereof. The kit is composed of a sample pad, a combined pad, a reaction pad, a water absorption pad and a PVC bottom plate, wherein a detection line T1 coated by an anti-CA125 antibody A2, a detection line T2 coated by an anti-HE4 antibody B2 and a quality control line C coated by goat anti-rat IgG are arranged on the reaction pad; by means of observing color development reaction of the detection lines, bedside quick screening diagnosis of early ovarian cancer can be achieved. The prepared nano selenium immunochromatography kit disclosed by the invention has the advantages of strong specificity,high sensitivity, good accuracy, convenience, quickness, small sample amount to be detected and suitability for screening, homelab and bedside quick detection of high-risk groups for ovarian cancer.
Owner:HENAN UNIVERSITY
Rapid detection kit for novel coronavirus antigen, preparation method and application of rapid detection kit
PendingCN112326966ASimple and fast operationSuitable for large-scale population screeningBiological material analysisAntigenReagent strip
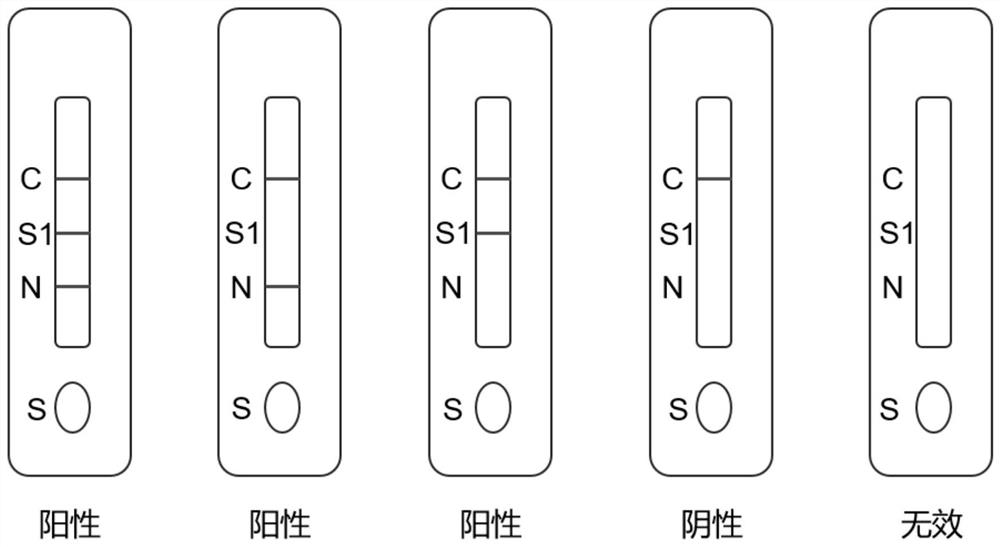
The invention discloses a rapid detection kit for a novel coronavirus antigen. The rapid detection kit comprises a kit body and a reagent strip placed in the kit body. The reagent strip comprises a bottom plate, a sample pad, a combination pad, a nitrocellulose membrane and a water absorption pad, wherein the sample pad, the combination pad, the nitrocellulose membrane and the water absorption padare sequentially overlapped end to end along the length direction of the bottom plate. The combination pad is coated with an anti-nucleocapsid protein monoclonal antibody and an anti-spinous glycoprotein monoclonal antibody which are labeled by colloidal gold; and the nitrocellulose membrane is coated with a nucleocapsid protein detection line N line formed by an anti-nucleocapsid protein monoclonal antibody, a spinous glycoprotein detection line S line formed by an anti-spinous glycoprotein monoclonal antibody, and a quality control line C line formed by a goat anti-mouse IgG antibody. The invention also discloses a preparation method and an application of the kit. The rapid detection kit prepared in the invention can be used for directly detecting the novel coronavirus, is simple and convenient to operate and short in detection time, and is suitable for large-scale population screening and on-site screening.
Owner:HANGZHOU YORK BIOTECH CO LTD
New corona-virus nucleic acid 10-in-1 test technology
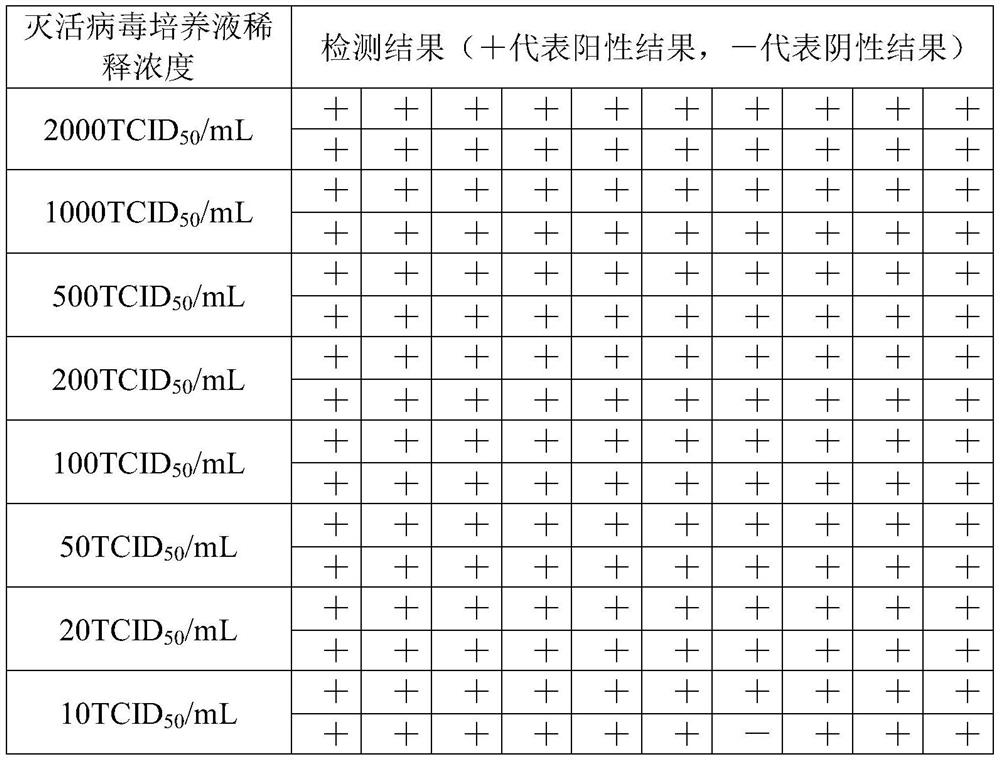
PendingCN111944927AReduce screening costsReduce medical collection consumablesMicrobiological testing/measurementSurgeryMicroorganismNucleic acid detection
The invention relates to the technical field of microorganisms, in particular to a new corona-virus nucleic acid 10-in-1 test (10-in-1 test) technology. The new corona-virus nucleic acid 10-in-1 testtechnology is a method for collecting 10 swabs collected from 10 persons in one collection tube and detecting, and the used materials comprise a virus collection tube and a collection swab. The 10-in-1 test technology provided by the invention is suitable for large-area general screening of people with low risk of new corona-virus infection, and can significantly reduce medical collection consumables required by large-scale population screening and reduce the nucleic acid detection cost of new corona-virus; according to the method, the low nucleic acid detection efficiency of a traditional collection and detection method is avoided, the SARS-CoV-2 infection can be effectively diagnosed as soon as possible, further diffusion of the SARS-CoV-2 infection can be controlled, and epidemic spreading can be prevented in time.
Owner:SHENGJING HOSPITAL OF CHINA MEDICAL UNIV
Primer set, chip and kit for genotyping detection of alpha thalassemia and beta thalassemia
PendingCN109112204ALow costReduce pollutionMicrobiological testing/measurementDNA/RNA fragmentationBeta thalassemiaPrenatal diagnosis
The present invention provides a primer set for detecting alpha thalassemia and beta thalassemia, and belongs to the field of molecular biology, wherein the primer set comprises an alpha primer set and a beta primer set. The invention further provides a chip capable of simultaneously detecting alpha thalassemia and beta thalassemia, wherein probes having sequences represented by SEQ ID NO:1-31 areimmobilized on the chip. Based on the primer set and the chip, the present invention further provides a kit capable of simultaneously detecting alpha thalassemia and beta thalassemia. The kit of theinvention has advantages of short detection time, low cost, convenient operation, closed tube operation and pollution reducing, and has great significance in the screening of thalassemia population, the genetic counseling and the prenatal diagnosis.
Owner:陈治中
Body fluid sulfydryl detection kit
ActiveCN103424402AImprove stabilityHigh speedMaterial analysis by observing effect on chemical indicatorMedicineChloride
The invention discloses a body fluid sulfydryl detection kit which comprises phosphotungstic acid, first buffer solution, mercuric chloride, first test paper and second test paper, wherein test paper becomes the first test paper after being soaked in phosphotungstic acid solution and then dried, other test paper becomes the second test paper after being soaked in mercuric chloride and the first buffer solution and then dried, the first test paper and the second test paper are adhered or close to each other in other ways to form compound test paper; body fluid is dripped on the first test paper, and the body fluid and reagents on the first test paper penetrate into the second test paper to detect the sulfydryl of the body fluid. All reagents components are compounded during the body fluid sulfydryl detection process, unstable chemical reagents separate the first buffer solution from phosphotungstic acid, which interact, through drying stabilization and mechanical structure design, so that the stability of the reagents at the normal temperature is improved greatly, any auxiliary reagents and devices are not required during the detection process, the speed is high, the operation is simple, and the large-scale population screening operation can be performed quickly.
Owner:FUJIAN MINGXI HAITIAN LANBO BIOTECH
Quantitative fluorescent multiplex PCR test kit for Epstein-Barr virus, and application thereof in nasopharyngeal carcinoma screening
InactiveCN102839222ATo achieve the purpose of high-throughput screening NPCAccurate detectionMicrobiological testing/measurementFluorescence/phosphorescenceMultiplexPromoter
The invention relates to a quantitative fluorescent multiplex PCR test kit for Epstein-Barr virus, and the kit comprises the following reagents: a, specific primer pairs for amplifying mutations of key target genes in the Epstein-Barr virus; and b, specific probes for the mutations of key target genes in the Epstein-Barr virus, wherein the mutations are P-thr, V-val and V-leu in the coding region of EBNA-1 gene, promoters Cp, Fp and Zp of the EBNA-1 gene, and XhoI-loss, 30bp Deletion and Ser 366 Thr of LMP1 gene. The test kit is advantageous in that: the aim of precise prediction of the risk of nasopharyngeal carcinoma in high risk population can be achieved; the main carcinogenic mutations can be well known for the combination of population screening and population intervention for nasopharyngeal carcinoma; and the kit has the advantages of high in sensitivity, fast in test, and low in cost, having broad market prospect.
Owner:广州市第十二人民医院
Process of micro circulating DNA by quantitative testing
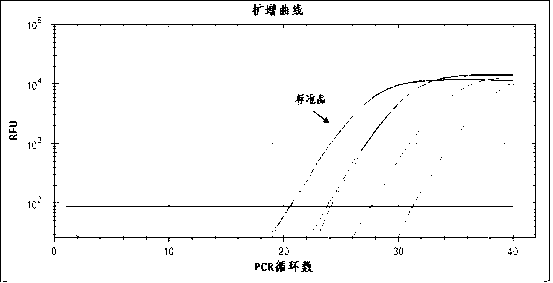
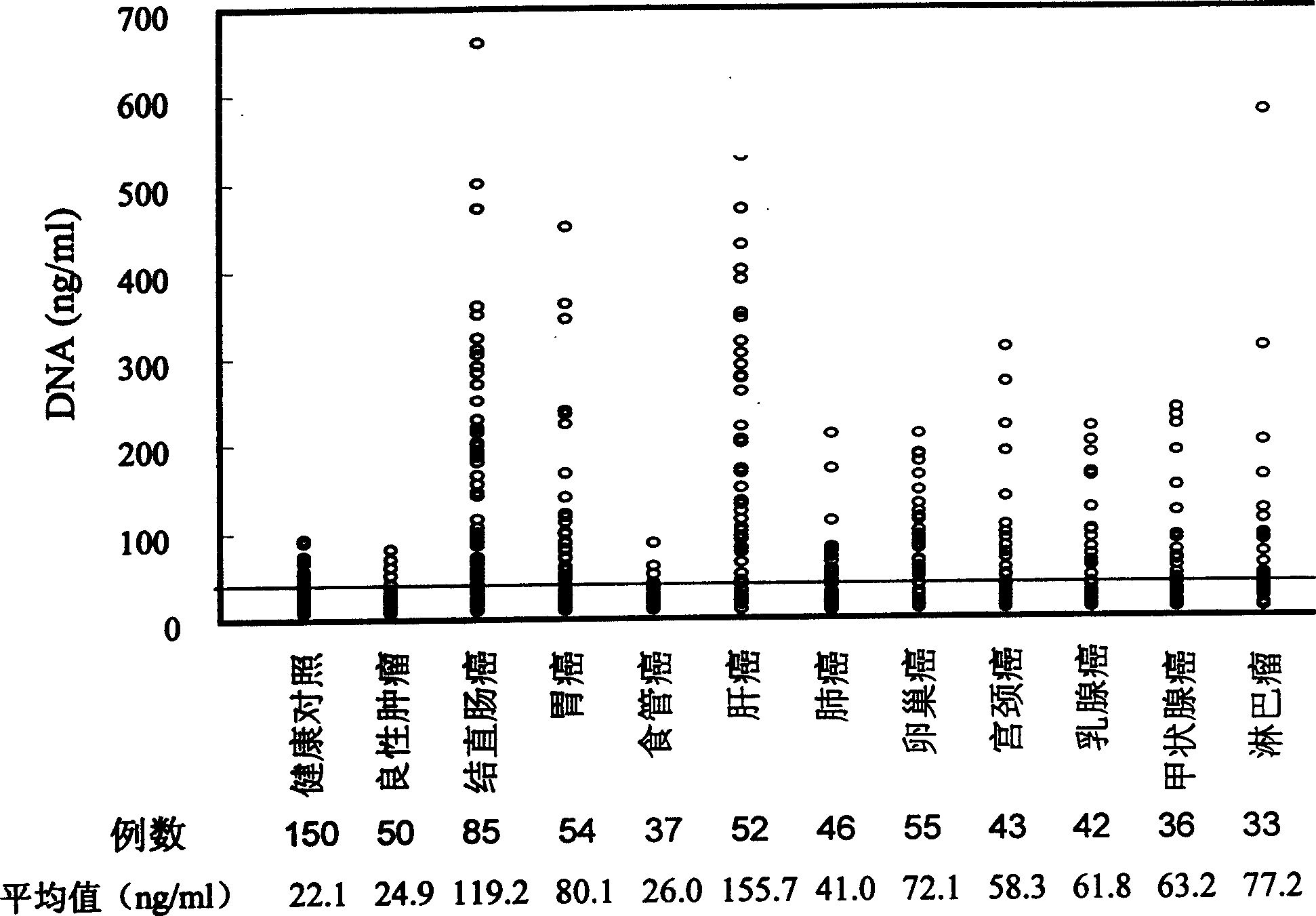
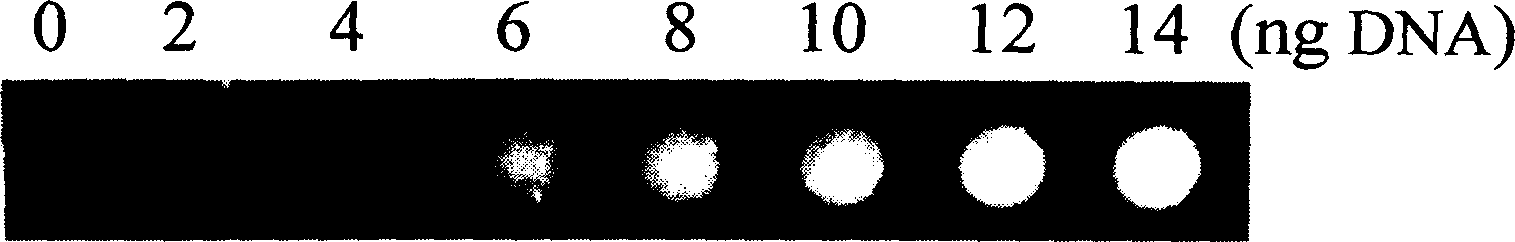
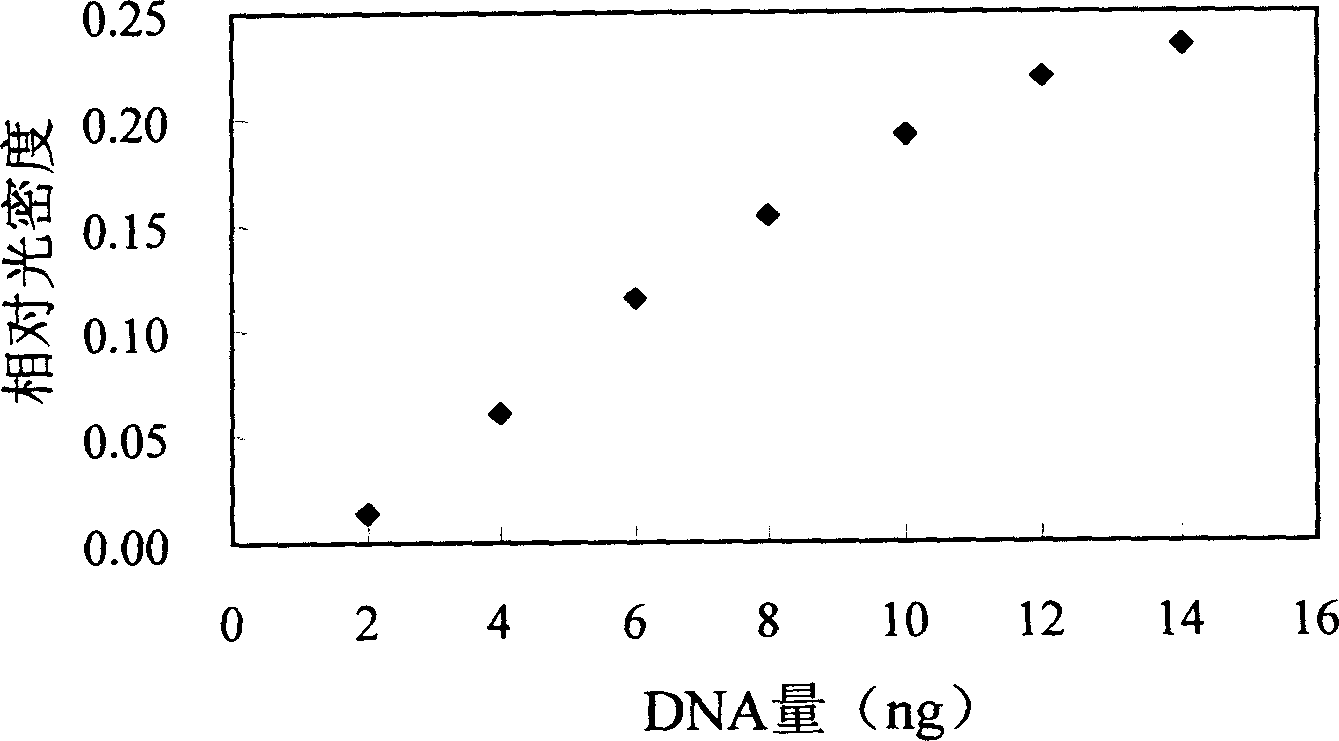
InactiveCN1560277AUniversal and convenientLow priceMicrobiological testing/measurementBiological testingCurative effectPopulation screening
The invention is a quantitative detecting technique of trace circular DNA based on fluorescent dye. It analyzes the strength of the fluorescence generated by combining the dye with purified DNA to determine the content of dissociative DNA in peripheral blood. It can accurately detect trace circular DNA down to a level of nano g, and has the characters of rapidness, simplicity and convenience, sensitivity, and accuracy. The determination of special people and tumor patient verifies that it has higher reliability, sensitivity and accuracy. It can be used in screening large-scale high-danger tumor people, early tumor diagnosis and dynamic observation of curative effect prognosis.
Owner:FUDAN UNIV
Typing detection kit for Candida albicans
ActiveCN104004851AImprove permeabilityEasy to operateMicrobiological testing/measurementNucleotideCandida famata
The invention discloses a typing detection kit for Candida albicans. The typing detection kit comprises a gene chip and various primers. The gene chip is provided with 13 different types of nucleotide sequence probes for the Candida albicans, a DNA sequence marked with biotin points and a DNA sequence with a coding beta-globulin part, wherein the probes are in the sequences of SEQ ID Nos: 1-13 or sequences complementary with the SEQ ID Nos: 1-13, the DNA sequence with the biotin points is SEQ ID No. 14, and the DNA sequence with the coding beta-globulin part is SEQ ID No. 15. The various primers are in the sequences of SEQ ID Nos: 16-21. By means of the typing detection kit for the Candida albicans, a typing joint detection platform for detecting the Candida albicans is provided, synchronous joint detection can be achieved, the detection of specificity can be enhanced, the cost can be lowered, the detection time can be shortened, and the typing detection kit has great significance for carrying out clinic early diagnosis and population screening for the Candida albicans.
Owner:上海凯普医学检验所有限公司 +2
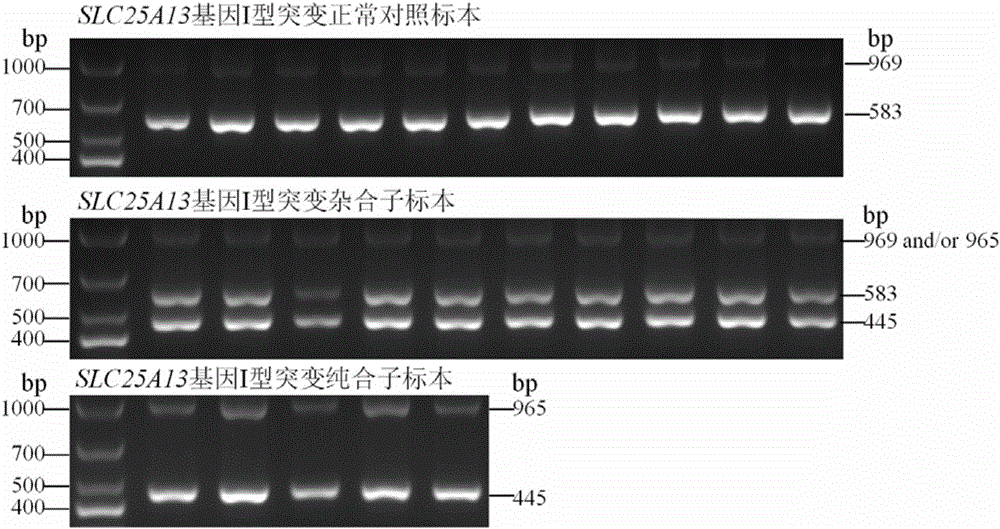
Citrin immunodeficiency disease pathogenic gene SLC25A13 high-frequency I-type mutation screening primers and kit
InactiveCN105803091AEasy to detectQuick checkMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyImmunodeficiency disease
The invention discloses Citrin immunodeficiency disease pathogenic gene SLC25A13 high-frequency I-type mutation screening primers and kit, belongs to the technical field of biology. Four newly designed primers are provided; a multi-PCR system depending on an ordinary rTaq enzyme is built; whether a sample is I mutation positive or negative or heterozygote can be identified through once PCR and analysis of an electrophoretogram; the primers and the kit are accurate, simple, fast and low in cost. By the screening primers and kit disclosed by the invention, DNA sequencing is not needed; the screening primers and kit are simple, fast and low in cost; the detection result is direct and reliable; the detection time and cost are superior to those of an existing method; the screening primers and kit can be finished by simple common equipment and are suitable for medical and test mechanisms of various regions for fast detection and large-scale population screening of the SLC25A13 high-frequency I-type mutation.
Owner:JINAN UNIVERSITY
Molecular markers for identifying PHYB wild type and mutant of rice phytochrome gene
ActiveCN106282345AValid identificationRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention discloses molecular markers for identifying the PHYB wild type and mutant of a rice phytochrome gene. The nucleotide sequences of the markers related to phyB wild type site detection and phyB mutant site detection are shown as Seq No.1-2 respectively. A primer PHYBF4 / PHYBR1 is used for carrying out PCR amplification, and an amplification product is subjected to BamH I digestion and electrophoretic analysis, wherein if the PCR product can not be digested by BamH I and is a 175bp DNA fragment, the PCR product is the wild type phyB, and if the PCR product can be digested by BamH I, and the digestion products are a 25bp DNA fragment and a 151bp DNA fragment, the PCR product is the mutant phyB1. The molecular markers have the advantages that amplification is easy and fast, and cost is low. The molecular markers can be used for effectively, rapidly and reliably identifying the phyB allelotype, and a powerful tool is provided for subsequent application of the phyB mutant in breeding population screening.
Owner:SHANDONG RICE RES INST
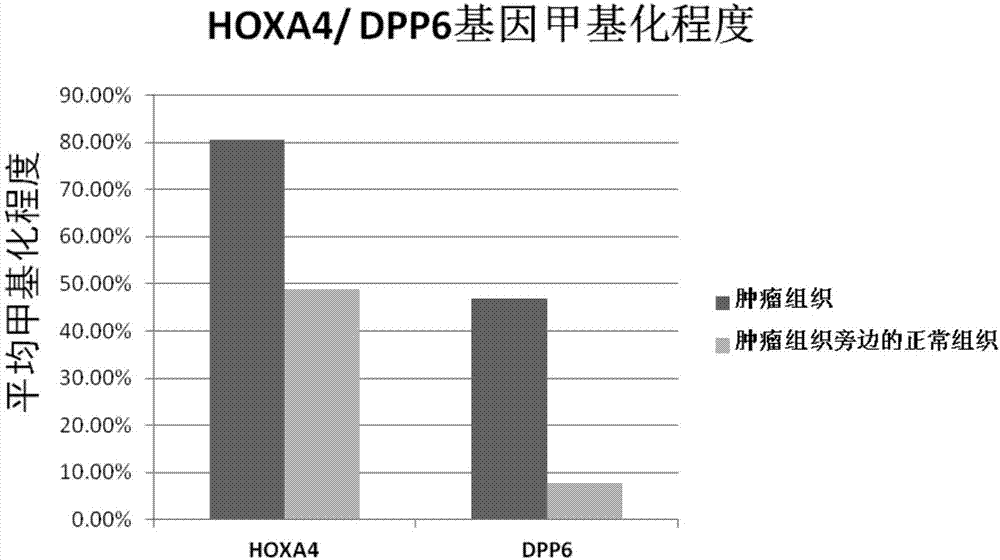
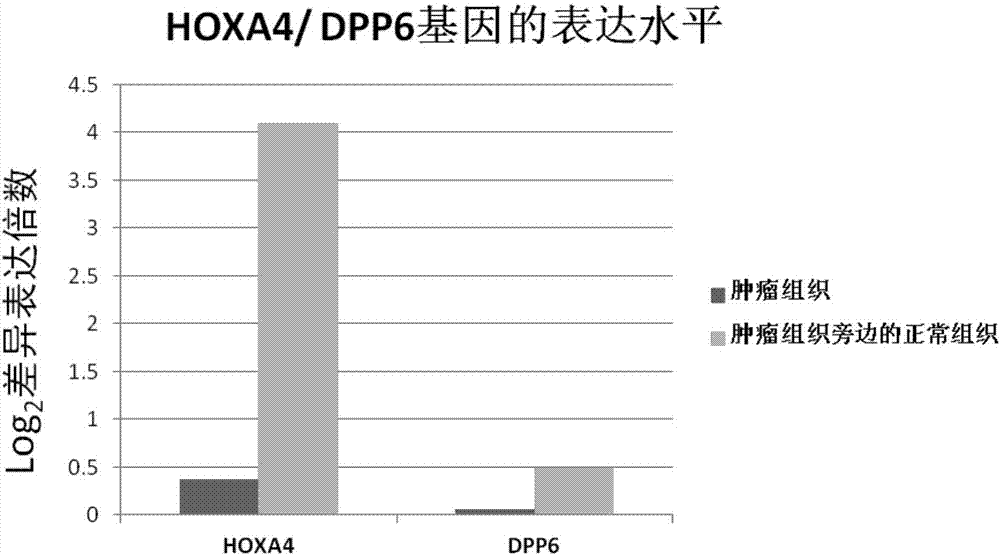
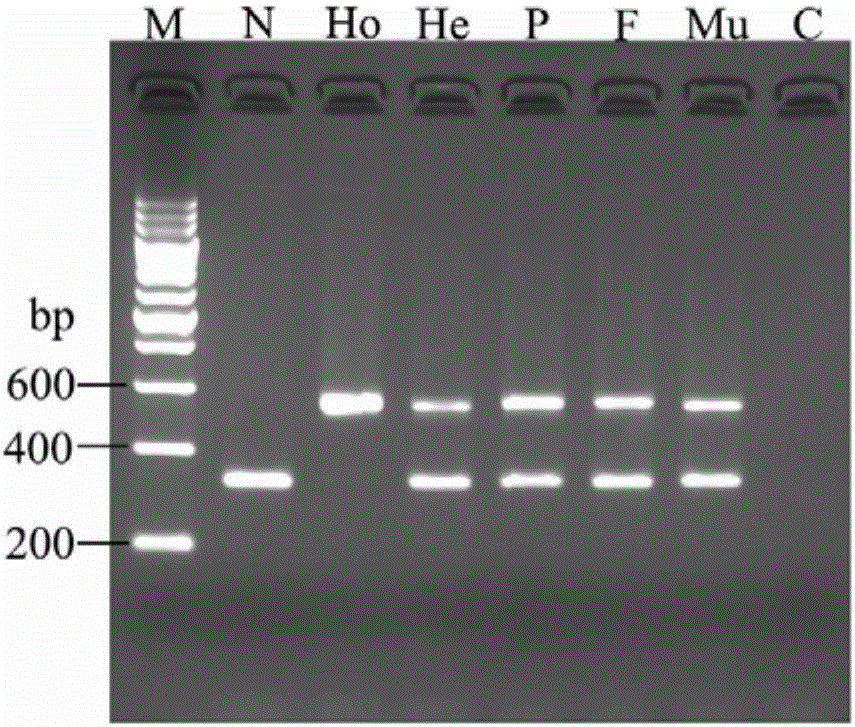
Kit for diagnosis and assessment of breast cancer, and detection and applications of methylated HOXA4/DPP6 gene
ActiveCN107988366AHigh expressionImprove accuracyMicrobiological testing/measurementTreatment targetsHOXA4
The invention relates to a kit for diagnosis and assessment of HOXA4 / DPP6 gene, and detection and applications of a methylated HOXA4 / DPP6 gene, and in particular to an HOXA4 / DPP6 gene, methylation detection of the HOXA4 / DPP6 gene, applications of the HOXA4 / DPP6 gene in preparing breast cancer diagnosis, detection and screening drugs and a kit for the applications. According to the invention, basedupon researches, it is discovered that a methylation degree of the HOXA4 / DPP6 gene in tumor tissues of a patient with the breast cancer is obviously higher than that of normal tissues beside the tumor tissues, and moreover, a methylation level is closely related to tumor progression; the invention provides a novel breast cancer diagnosis and treatment target and provides a kit corresponding to the breast cancer diagnosis and treatment target; therefore, a novel method is provided for detecting the progression of the breast cancer and for population screening and breast cancer diagnosis, and an important clinical application value is achieved.
Owner:SHENZHEN CITY BAOAN DISTRICT MATERNAL & CHILD HEALTH HOSPITAL
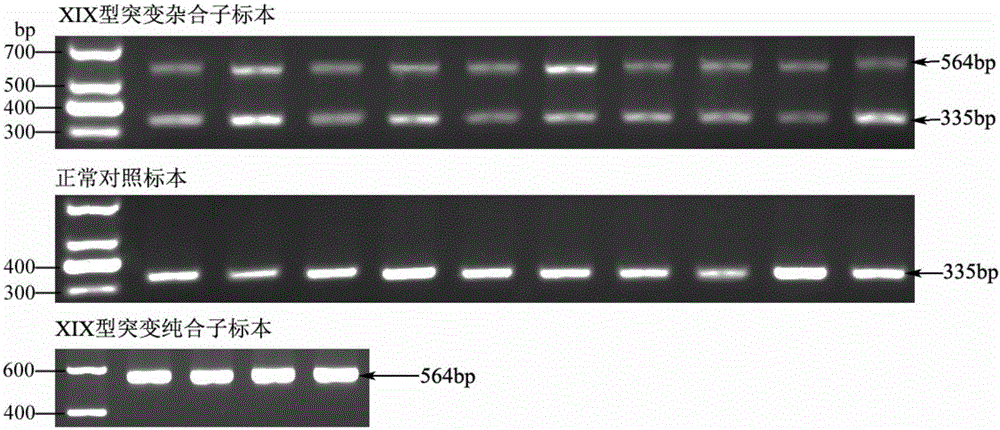
High-frequency XIX type mutation screening primer and high-frequency XIX type mutation screening kit for disease-causing gene SLC25A13 of Citrin deficiency (CD)
InactiveCN105695592AEasy to detectQuick checkMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMutation screening
The invention discloses a Citrin deficiency disease-causing gene SLC25A13 high-frequency type XIX mutation screening primer and a kit, which belong to the field of biotechnology. The present invention provides three newly designed primers, establishes a multiplex PCR system relying on common Taq enzymes, and can identify whether a sample is XIX mutation negative, positive or heterozygous through one PCR and analysis of the electrophoretic pattern, which is accurate, simple, fast and low cost. The screening primers and kits of the present invention do not require DNA sequencing, are simple, fast, and low-cost, and the detection results are direct and reliable. The detection time and cost are significantly better than existing methods, and can be completed with simple and common equipment, suitable for Rapid detection and large-scale population screening of high-frequency XIX mutations of the SLC25A13 gene by medical and testing institutions around the world.
Owner:JINAN UNIVERSITY
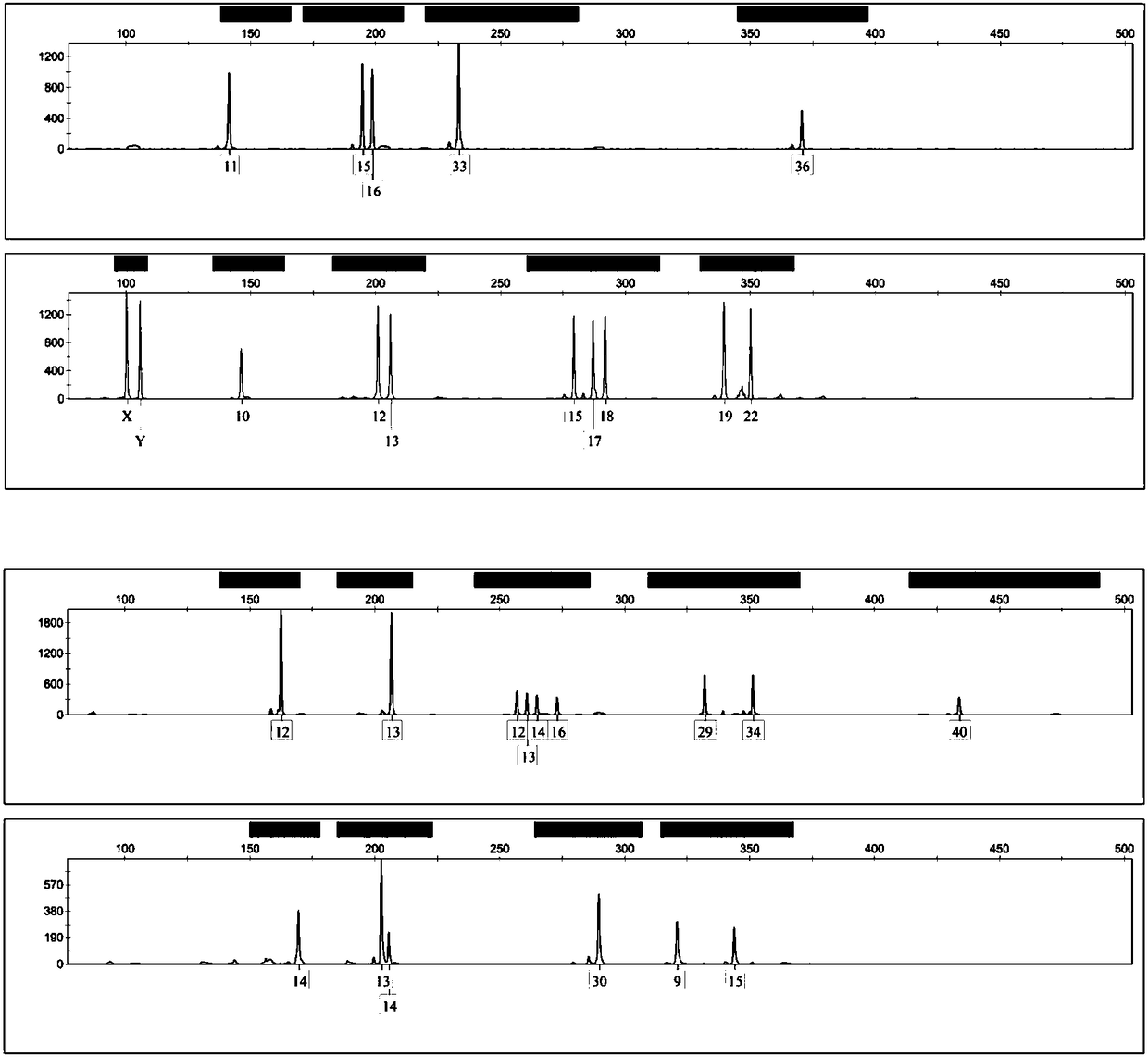
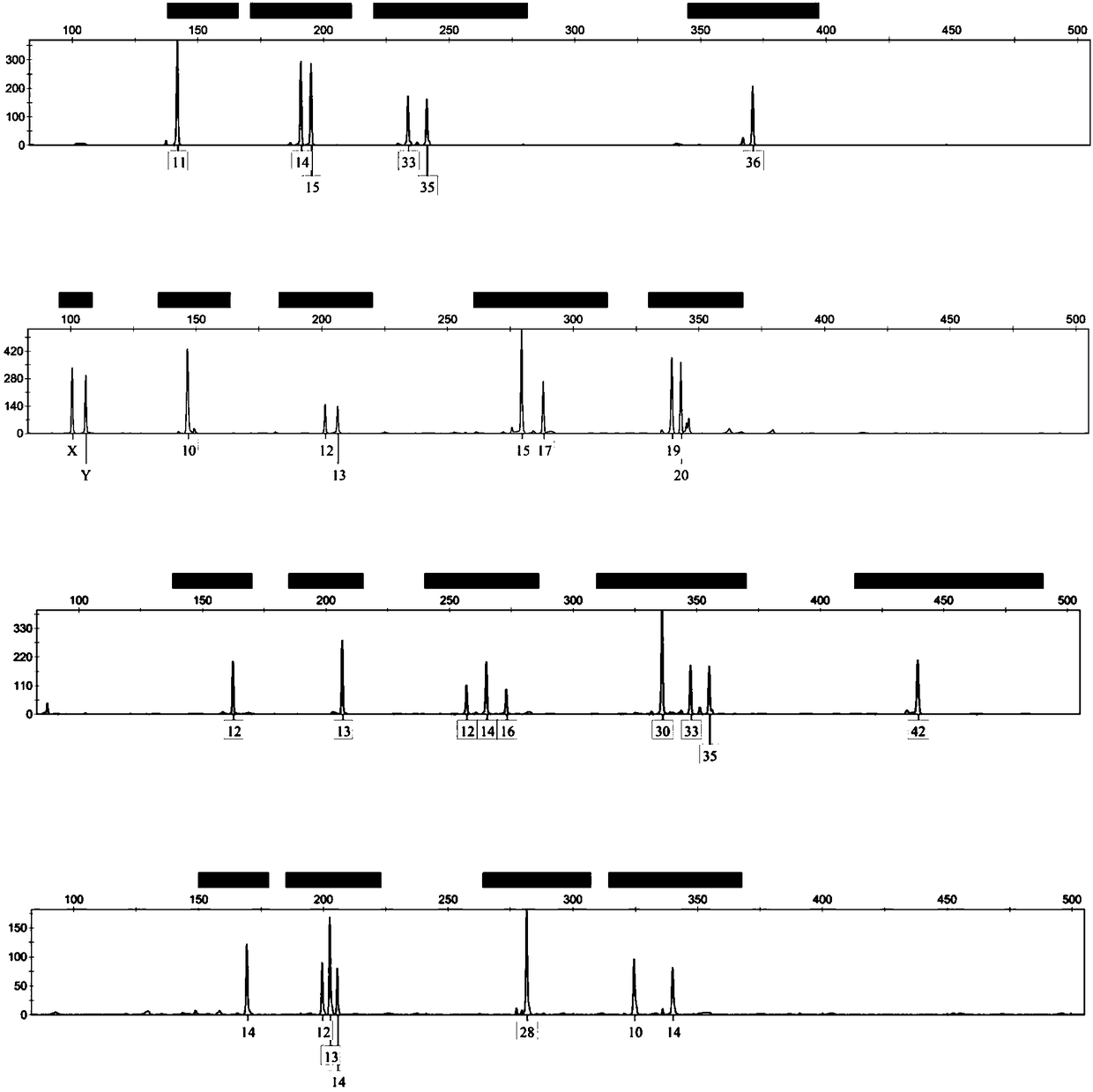
Composite amplification system based on Y-STR locus and specific primer combination thereof
ActiveCN108866201AMeet the inspection requirementsEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationStr typingAgricultural science
The invention discloses a composite amplification system based on a Y-STR locus and a specific primer combination thereof. The primer combination provided by the invention is formed by 30 kinds of DNAmolecules shown as sequence 1 to sequence 30 in a sequence table. The primer combination is used for building the composite amplification system based on the Y-STR locus; then, male individual STR typing is performed; the detected polymorphism is high; the population screening and individual recognition capability of a Y chromosome genetic marker can be enhanced; particularly important identification values are realized on the distant relatives of the same family. Therefore when the primer combination provided by the invention is used for sample screening, the cost is low; the efficiency is high; important application values are realized.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Molecular marker of myopia-related susceptibility genes as well as detection primer group and application thereof
ActiveCN111893179AMany categoriesThe number of detected gene loci is largeMicrobiological testing/measurementDNA/RNA fragmentationHuman DNA sequencingGenetic risk
The invention is applicable to the fields of biotechnology and ophthalmic health, and provides a molecular marker of a myopia-related susceptibility gene as well as a detection primer group and application of the molecular marker. The detection primer group of the molecular marker comprises multiple PCR amplification upstream and downstream primer pairs of 37 SNP loci of 24 high myopia and pathological myopia-related susceptibility genes of human genome DNA, and a single base extension primer. According to the invention, sample nucleic acid mass spectrum detection can be carried out by using amultiplex PCR amplification technology, the 37 SNP loci can be detected at one time by double reaction holes, through correlation analysis of highly myopia susceptibility genes and genetic risks, a highly myopia genetic risk level assessment method special for Chinese population is provided by using a highly myopia genetic risk assessment system, and the method has the advantages of high detection flux, high efficiency, high accuracy, low cost and the like, and is suitable for large-scale population screening.
Owner:陕西九州医学检验有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com