Method and kit for diagnosing bladder cancer with urine
A technology of bladder cancer and detection kit, applied in the field of biomedical detection, can solve problems such as small sample size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] Example 1. High-throughput methylation profiling at the genome level
[0107] The present invention conducts high-throughput genomic analysis on two cases of normal human bladder mucosa (obtained from Urology Department of Zhongshan Hospital) and two cases of bladder cancer cell lines (5637 (ATCC: HTB-9), T24 (ATCC-4)). Establishment of methylation profiles.
[0108] Extract the above-mentioned bladder cancer cells and normal tissues, and use MBD (methylation binding protein domain) affinity chromatography column to enrich the medium and low methylated DNA fragments step by step; step 2: the collected hypermethylated DNA fragments Add SOLEXA adapters and send to Shanghai Bohao Biotechnology Co., Ltd. for SOLEXA high-throughput sequencing. The bladder cancer tumor cell group and the normal bladder tissue group obtained 3 million reads respectively. After bioinformatics analysis, genome mapping, and the positioning information was uploaded to UCSC to establish a database...
Embodiment 2
[0109] Example 2, preliminary screening of methylation difference
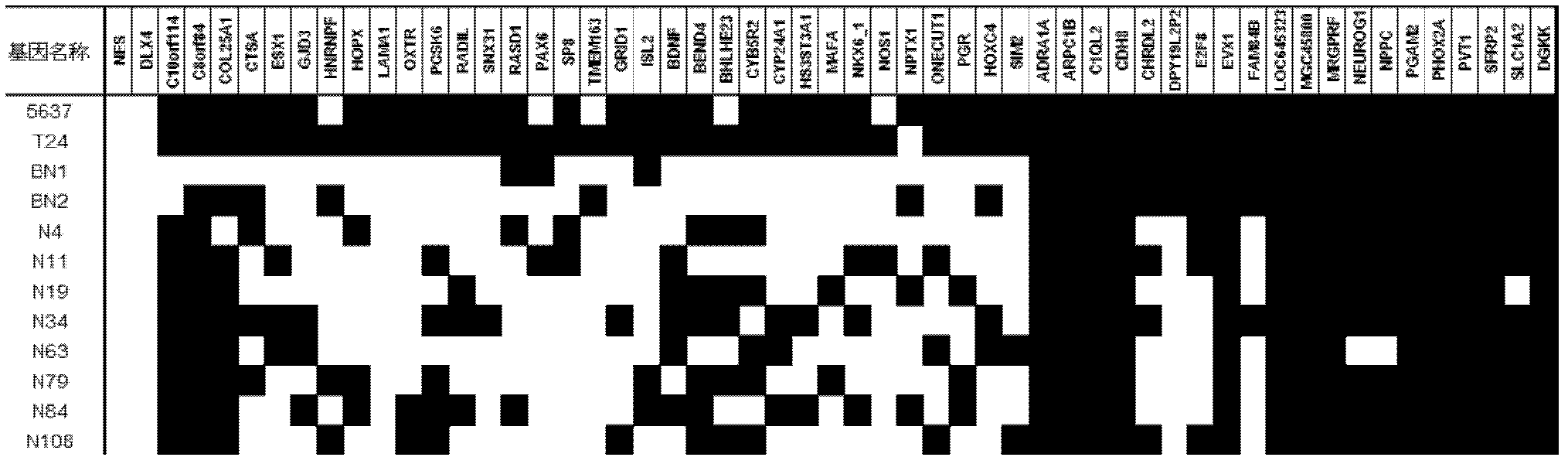
[0110]Further screened from the above 1627 highly methylated regions associated with gene promoters in tumor cell lines to obtain the promoter-related regions of the top 104 genes with the largest difference in methylation value (sorted according to P value, select the region related to the normal bladder The top 104 genes with the highest degree of methylation compared to tissues). Including: NES, DLX4, C10orf114, C8orf84, COL25A1, CTSA, ESX1, GJD3, HNRNPF, HOPX, LAMA1, OXTR, PCSK6, RADIL, SNX31, RASD1, PAX6, SP8, TMEM163, GRID1, ISL2, BDNF, BEND4, BHLHE23, CYB5R2, CYP24A1, HS3ST3A1, MAFA, NKX6_1, NOS1, NPTX1, ONECUT1, PGR, HOXC4, SIM2, ADRA1A, ARPC1B, C1QL2, CDH8, CHRDL2, DPY19L2P2, E2F8, EVX1, FAM84B, LOC645323, MGC45800, MRGRPCRF, PGAM2, PHOX2A, PVT1, SFRP2, SLC1A2, DGKK, ACTA1, CCND1, CYP26B1, FGF3, FOXD3, LHX9, NEUROG2, OPRK1, PADI2, SLC6A4, BARHL1, CDO1, TUBB2B, IHH, TBX20, SLC46A2, PDZK1P1, FEZF2, B ...
Embodiment 3
[0119] Example 3, Further Screening for Differences in Methylation
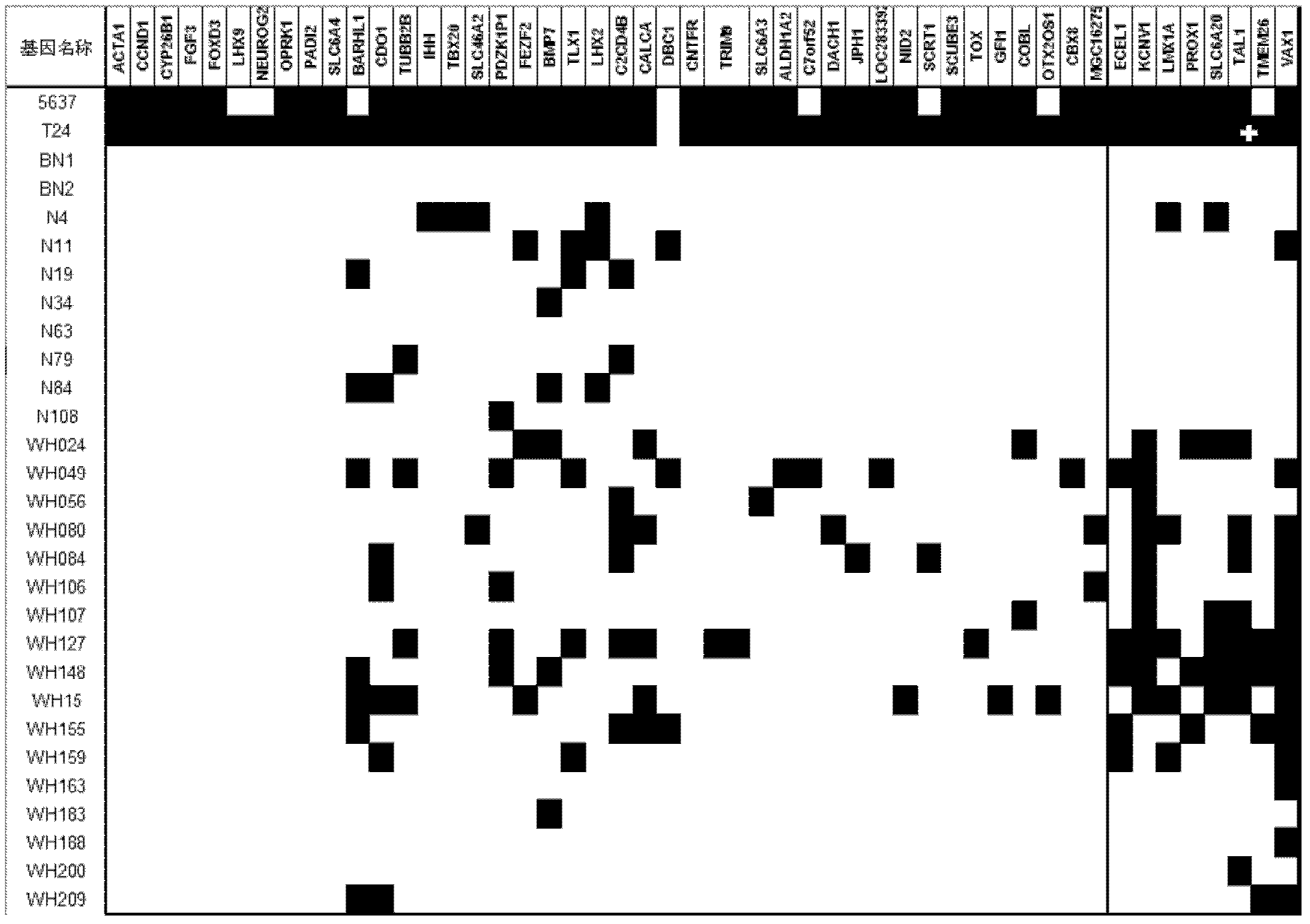
[0120] The tested samples were bladder cancer cell lines 5637 (ATCC: HTB-9) and T24 (ATCC-4), 2 cases of normal bladder tissue, 8 cases of normal human urine samples (the urine sediment was obtained), bladder cancer There were 17 urine samples from patients. The urine samples were randomly selected, and the selection criteria were that the amount of genomic DNA was large enough to deal with multi-gene screening.
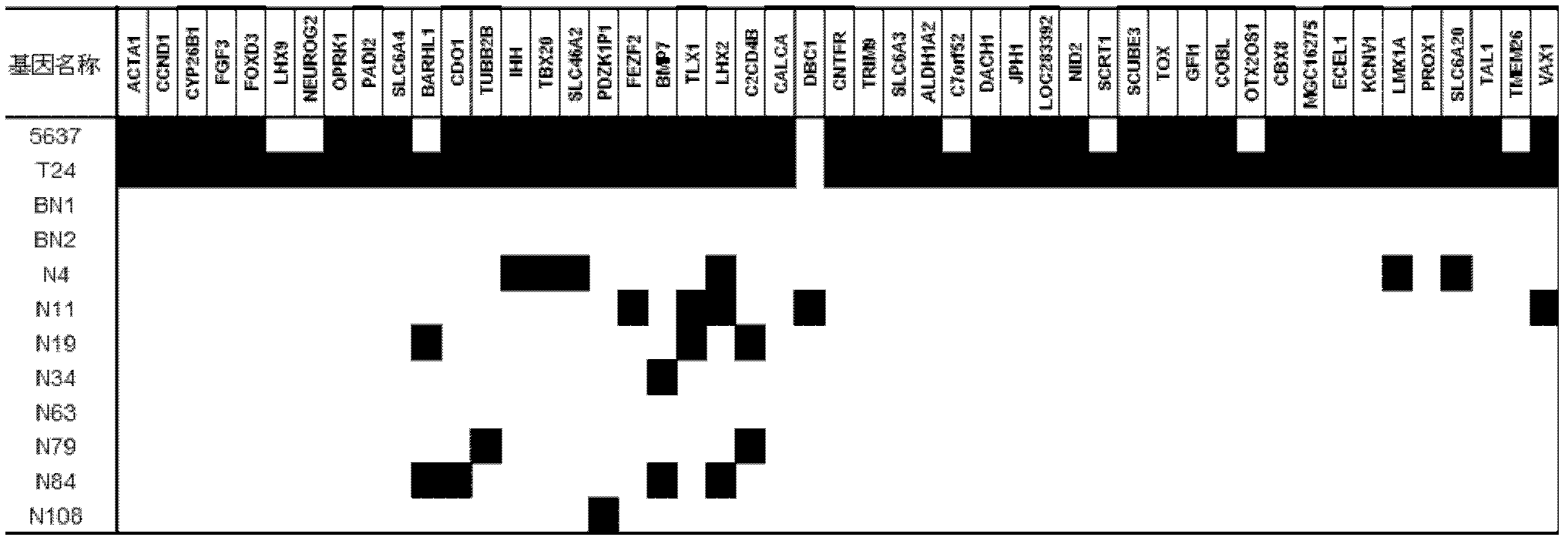
[0121] The specific regions of the 49 gene promoters screened in Example 2 were detected by methylation-specific PCR (MSP). The chemical modification of the sample with ammonium bisulfite and the methylation-specific PCR analysis method are the same as in Example 2.
[0122] see results image 3 . As a result, the inventors set the screening criteria as 4, that is, there were at least 4 cases of methylation in the urine samples of 17 bladder cancer patients, and the targets with a methylation rat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com