Single nucleotide polymorphism (SNP) molecular markers and method for pork DNA tracing by high resolution melting (HRM) method
A technology of molecular markers and pork, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, biochemical equipment and methods, etc., can solve the problems of long detection cycle, complex operation and high cost of pork DNA traceability methods, and achieve low detection cost Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
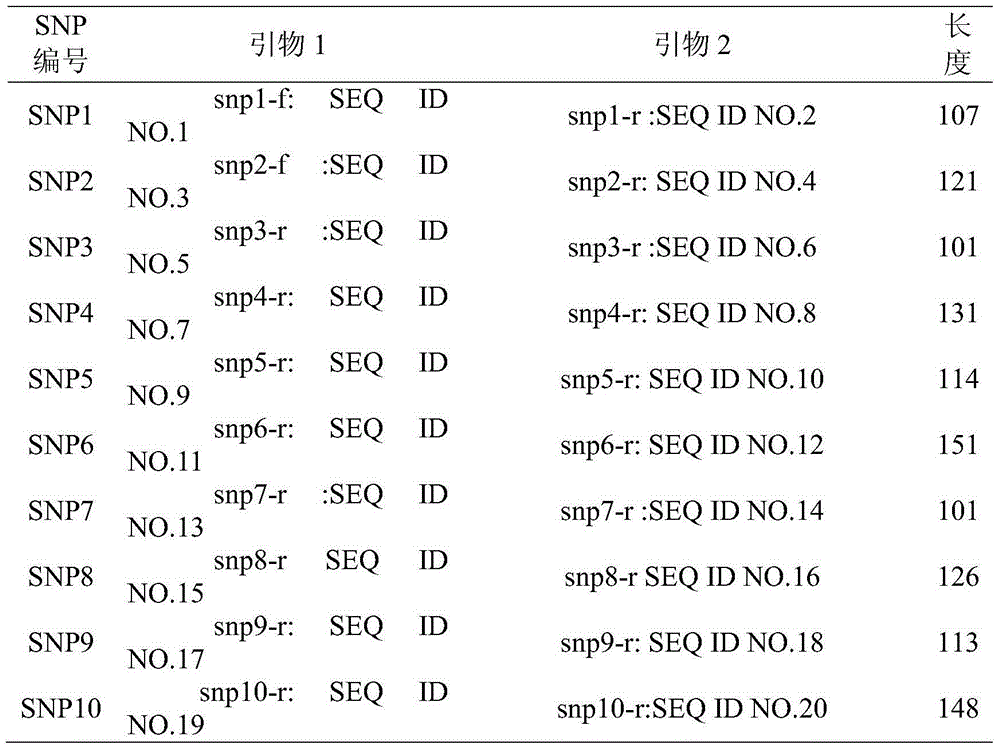
[0065] (1) Selection of SNPs sites
[0066] Select SNPs sites from NCBI to ensure that ① the distance between any adjacent SNPs is ≥ 1Mb, and that the SNPs are independent of each other and that there is no linkage; ② there is no relationship between any SNPs and other molecular markers; ③ the flanking sequences of SNPs are not Any variation exists. The SNPs selected in this study are listed in Table 1.
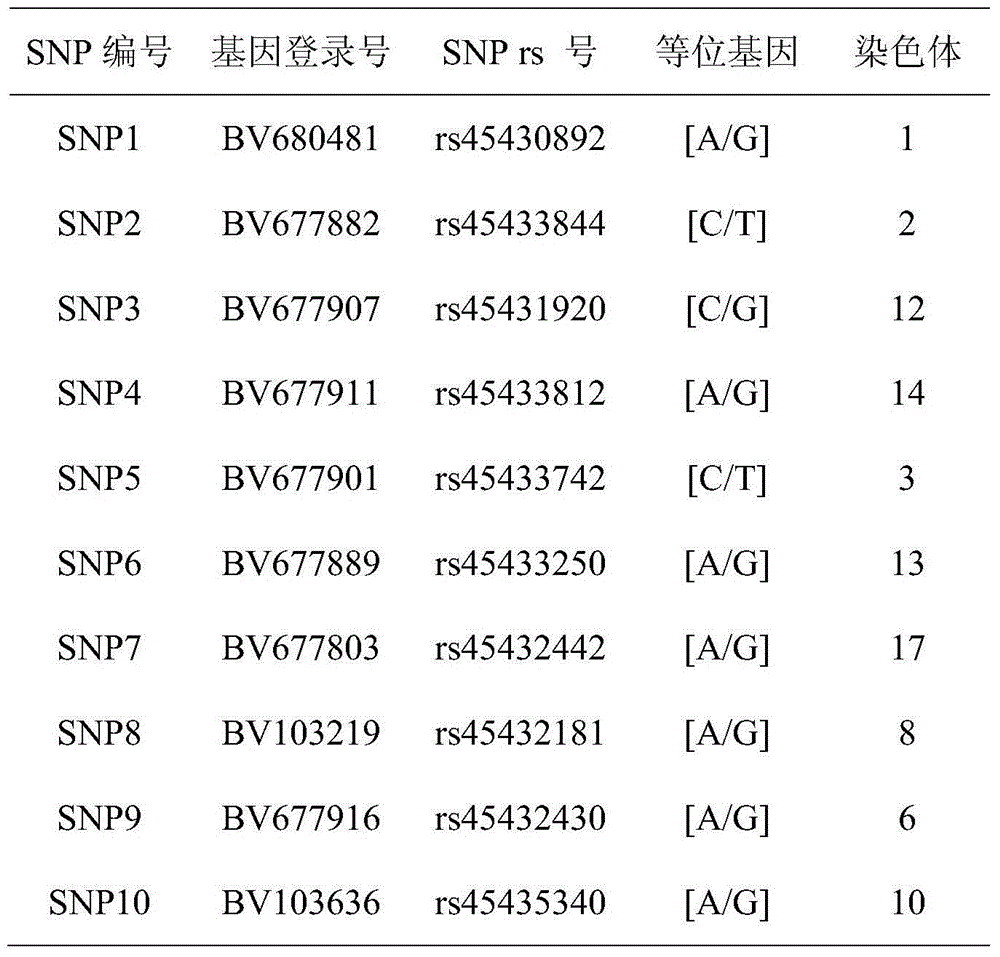
[0067] Table 1 SNP site related information
[0068]
[0069] As shown in Table 1, among the selected 10 SNP sites, SNP1 is located on chromosome 1 of the pig genome, SNP2 is located on chromosome 2 of the pig genome, SNP3 is located on chromosome 12 of the pig genome, SNP4 is located on chromosome 14 of the pig genome, and SNP5 is located on chromosome 14 of the pig genome On chromosome 3, SNP6 is located on chromosome 13 of the pig genome, SNP7 is located on chromosome 17 of the pig genome, SNP8 is located on chromosome 8 of the pig genome, SNP9 is located on chromosom...
Embodiment 2
[0102] 1) Pig DNA sample preparation
[0103] Select the fresh pork individuals of No. 1, 2, 5, 6, 30, 31, 40, 48, 49, and No. 53 in the above-mentioned 299 samples. When the pig muscle tissue and blood were collected in Example 1, 10 pigs were selected here. Cut 10 pig skin samples with pig hair from the individual, record the individual number of each sample, mark the samples, and shuffle the samples to be tested.
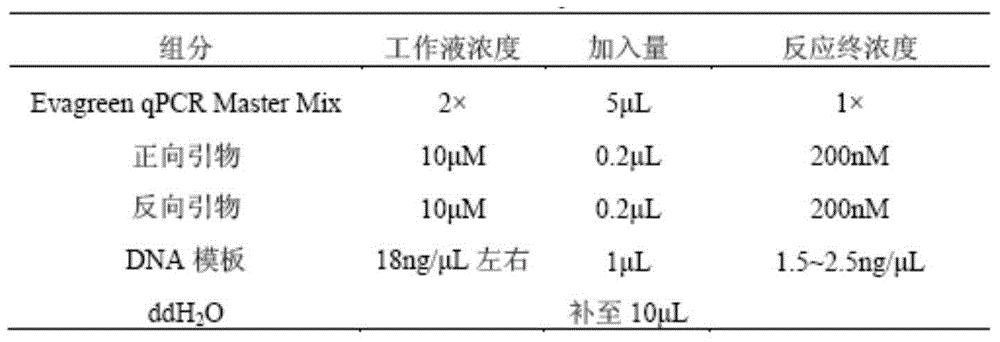
[0104] Extract 15 pig hairs with complete hair follicles from the pigskin of each sample, cut as short as possible only the hair follicles, put them into a clean 1.5ml centrifuge tube, add 200ul of DNA lysate and 10ul of proteinase K, and mix well . Digest at 56°C for 2 hours, during which the samples were flicked with fingers to speed up the digestion process. Add 60ul of 5mol / L NaCl and mix thoroughly; add 260ul of saturated phenol / chloroform / isoamyl alcohol (25:24:1), mix by inversion for 5min to fully denature the protein, centrifuge at 1000r / min for 5min, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com