Molecular marker subjected to co-segregation with pea powdery mildew resistance allele er1-6 and application thereof
A technology of molecular markers and alleles, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems that the effectiveness of functional markers has not been verified, and achieve the advantages of simple operation and reduced yield loss Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Identification of the er1 candidate gene PsMLO1cDNA sequence of the resistance resource and identification of the new allele er1-6
[0038] The RNAprep plant total RNA extraction kit (spin column type, purchased from Tiangen Company) was used to extract the total RNA of 15 pea disease-resistant resources and the susceptible control variety Baying No. 6. The specific operation method is as follows:
[0039] 1) Take about 100 mg of young pea leaves and quickly grind them into powder in liquid nitrogen, add 450 μL of RL (before use, add β-mercaptoethanol to a final concentration of 1%), and vortex vigorously to mix; 2) Transfer all solutions to On the filter column CS (the filter column CS is placed in the collection tube), centrifuge at 12000rpm for 5min, carefully draw the supernatant in the collection tube into the RNase-free centrifuge tube; 3) Slowly add 0.5 times the volume of anhydrous Ethanol, mix well, transfer the obtained solution and the precipitate ...
Embodiment 2
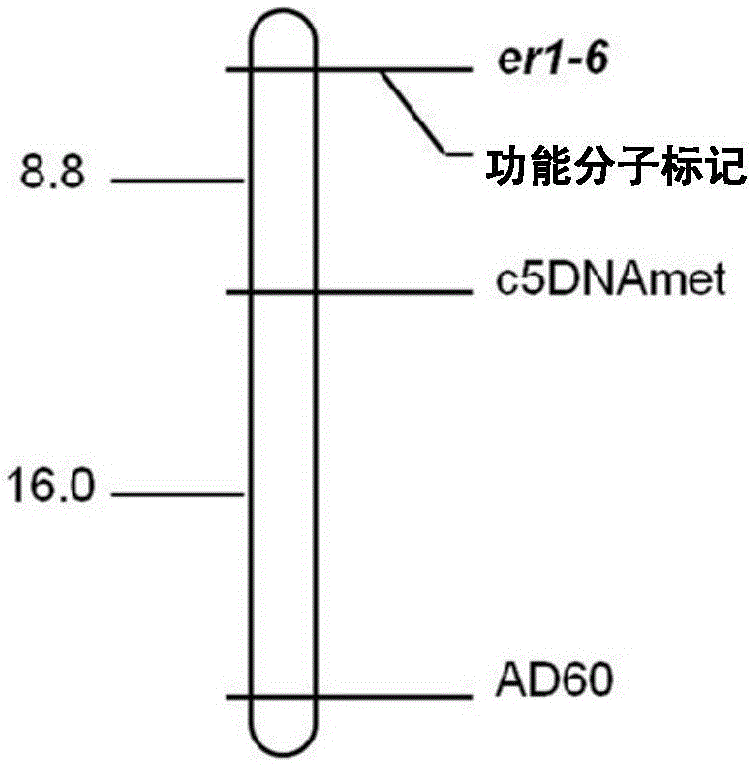
[0046] Example 2 Development of molecular markers co-segregated with pea powdery mildew resistance allele er1-6
[0047] According to the results of the gene PsMLO1 cDNA sequence comparison between the pea powdery mildew resistant variety G0001778 and the susceptible variety Bawan 6 and the wild-type PsMLO1 sequence, it was found that G0001778 contained a new allele er1-6, which was opened in the PsMLO1 sequence. A single base substitution (T→C) occurred at the 1121st position of the reading frame, and PrimerPremier5.0 software was used to design upstream and downstream primers on both sides of the SNP mutation site to develop functional molecular markers.
[0048] Upstream primer F: 5'-CTGGAGATCACCTTTTCTGGTT-3'
[0049] Downstream primer R: 5'-CATGTACAAACACACATACACACG-3'
[0050] The upstream and downstream primers of the functional marker are respectively located on the 11th exon and the 11th intron of the PsMLO1 gene. Primers were synthesized by Sangon Bioengineering (Sha...
Embodiment 3
[0055] Example 3 Application of molecular markers co-segregated with pea powdery mildew resistance allele er1-6
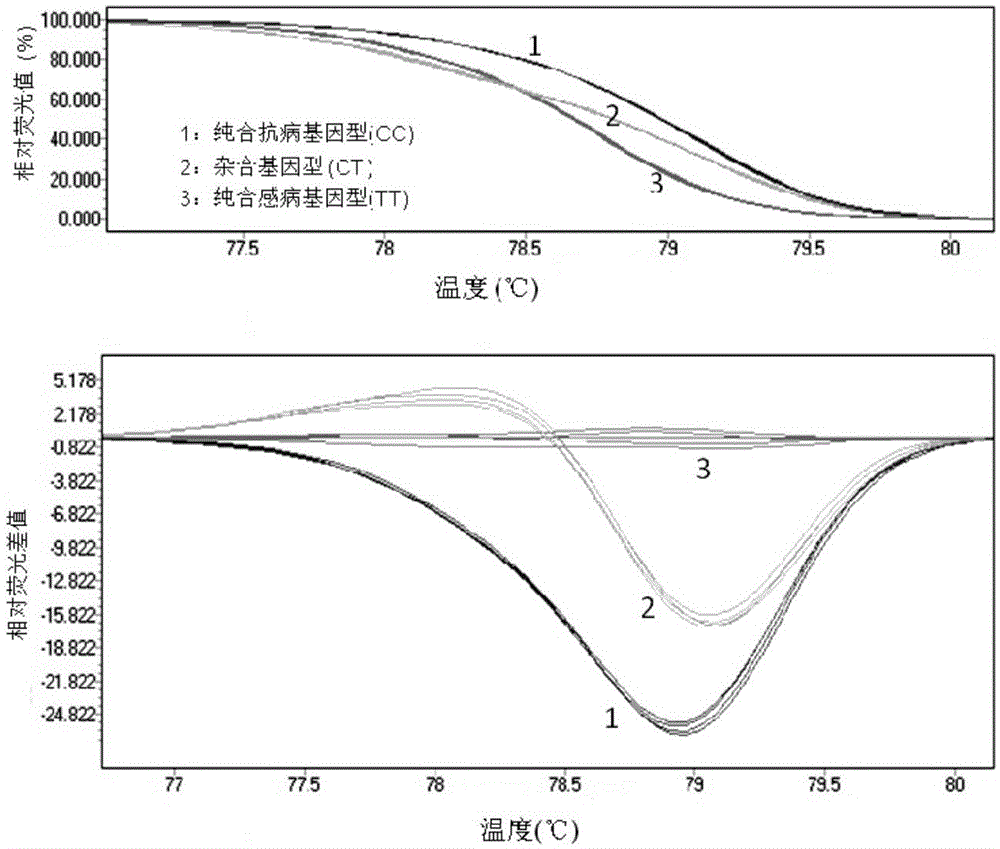
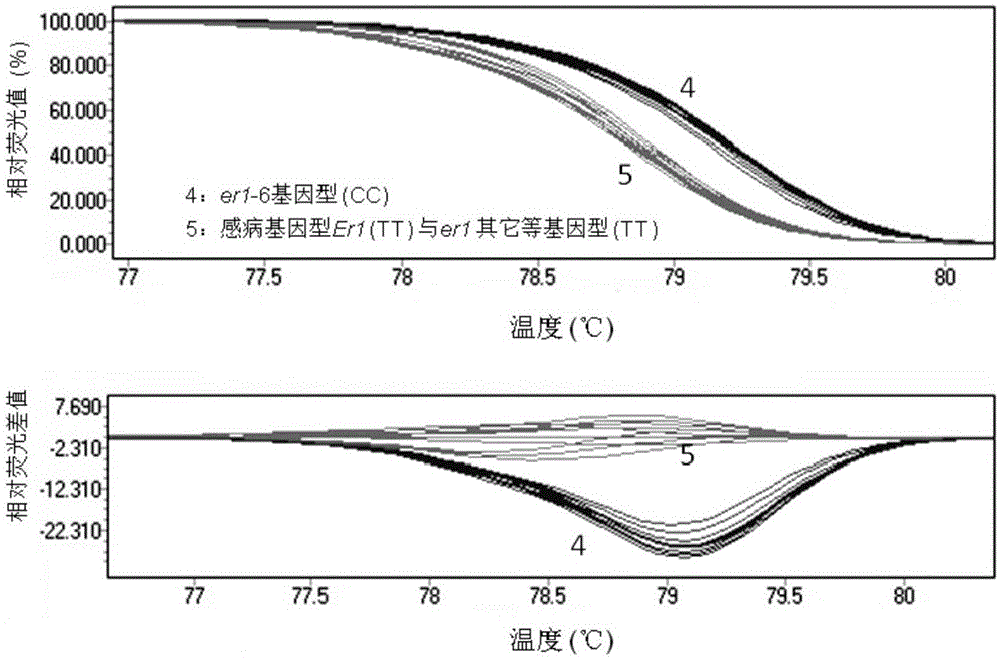
[0056]The developed er1-6 functional molecular markers were applied and functionally verified on pea variety resources. The results showed that the molecular marker amplified a 158bp target band in all detected disease-resistant and susceptible pea resources. Through HRM analysis technology, the pea resources containing the disease-resistant allele er1-6 (G0001752, G0001763, G0001764, G0001768, G0001778, identified by the PsMLO1cDNA sequence, all have a single base mutation T→C at 1121bp of the PsMLO1cDNA sequence) formed The same melting curve ( image 3 ). Disease-resistant resources containing other er1 alleles er1-1 (Tara), er1-2 (X9002 and G0005576), er1-4 (YI), and susceptible resources (Bawan 6, Longwan 1, G0001747, G0003839, G0003840) all form a melting curve which is obviously different from that of er1-6 ( image 3 ). The results showed that the mark...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com