Primer group and kit for simultaneously detecting four diarrhea viruses
A technology for detecting kits and diarrhea viruses, which is applied in the direction of microorganisms, recombinant DNA technology, and methods based on microorganisms. It can solve the problems of no HRMA analysis technology and no simultaneous detection, and achieve high sensitivity and specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Design of specific primers
[0043]First, download as many genomic DNA sequences of rotavirus, astrovirus, norovirus and saruvirus as possible from the NCBI gene bank in the United States, and then use the molecular biology software Bioedit to perform homologous comparison analysis on the downloaded sequences to find Highly homologous conserved sequences were used as candidate regions for primer and probe design, and primer design was performed in conjunction with the software PrimerSelect. The core design idea is: comprehensively consider the specificity and versatility of the detection of diarrhea virus (use degenerate bases at the viral nucleic acid mutation sites), and the universal principles that should be followed in primer design (such as Tm value, 3' terminal free energy , GC content, avoidance of internal secondary structure and dimer formation, etc.), and the interaction between primers during multiple amplification reactions, etc. After the desig...
Embodiment 2
[0049] Embodiment 2: The composition and detection method of the kit for detecting four kinds of diarrhea viruses based on HRMA technology
[0050] 1. The composition of the kit (stored at -20°C)
[0051] (1) 2X amplification reaction solution: its components are: 100mM Tris-HCl (pH8.3), 100mM KCl, 20mM MgCl2, 20mM DTT, 1mM Spermidine, 0.1% Tween-20, 0.2mg / L BSA, 0.4mM dNTP, 2.4 μM EvaGreen, 1.2 μM primer F1, 1.2 μM primer R1, 0.4 μM primer F2, 0.4 μM primer R2, 1.6 μM primer F3, 1.6 μM primer R3, 1.6 μM primer F4, 1.6 μM primer R4; wherein primer F1 is The sequence number is the nucleotide sequence shown in SEQ ID NO: 1, the primer R1 is the nucleotide sequence shown in SEQ ID NO: 2, and the primer F2 is the nucleic acid sequence shown in SEQ ID NO: 3. Nucleotide sequence, primer R2 is the nucleotide sequence shown in SEQ ID NO: 4, primer F3 is the nucleotide sequence shown in SEQ ID NO: 5, primer R3 is the sequence number shown in SEQ ID The nucleotide sequence shown in NO...
Embodiment 3
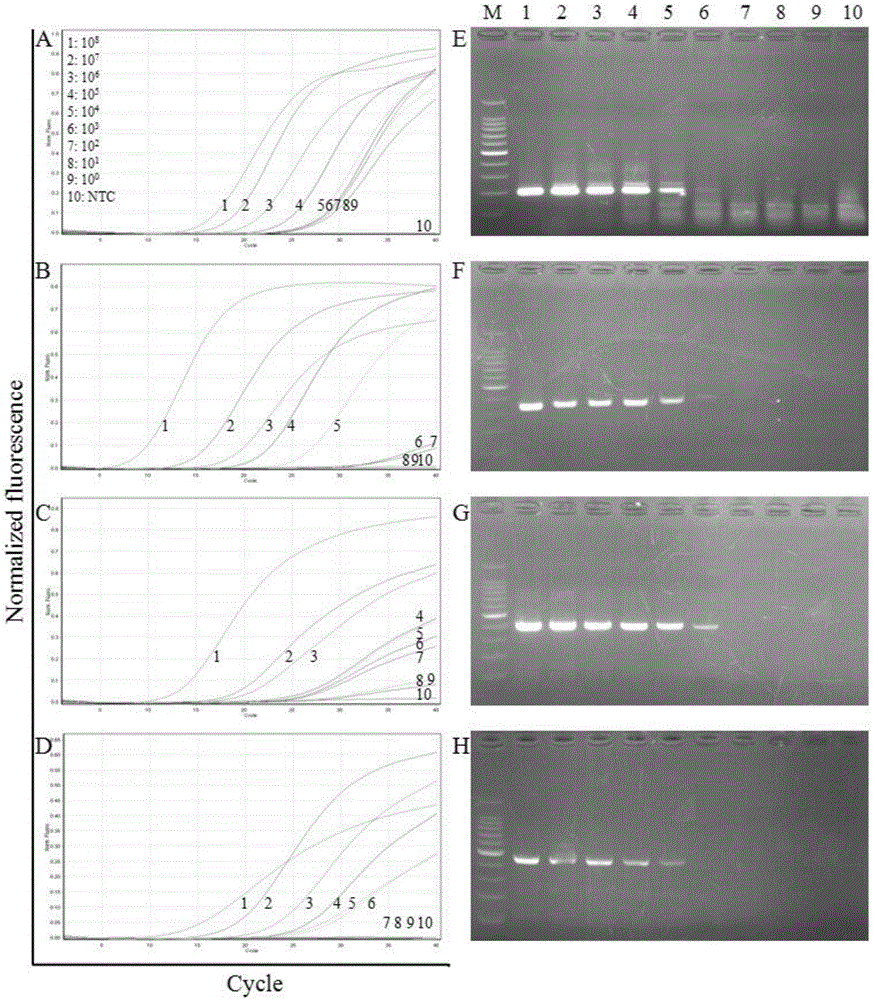
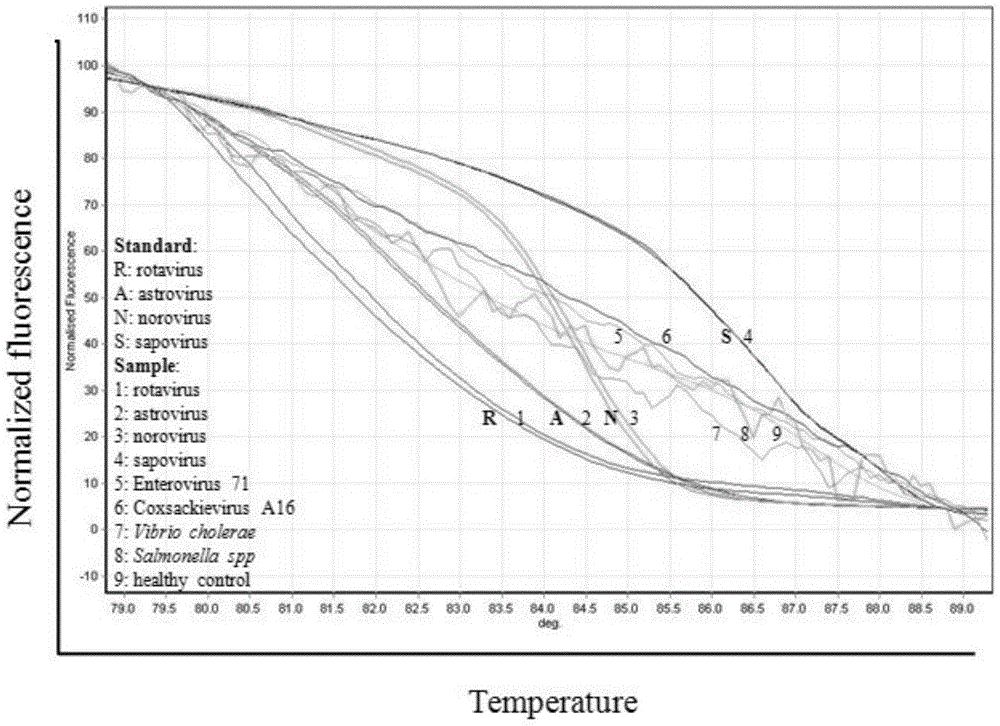
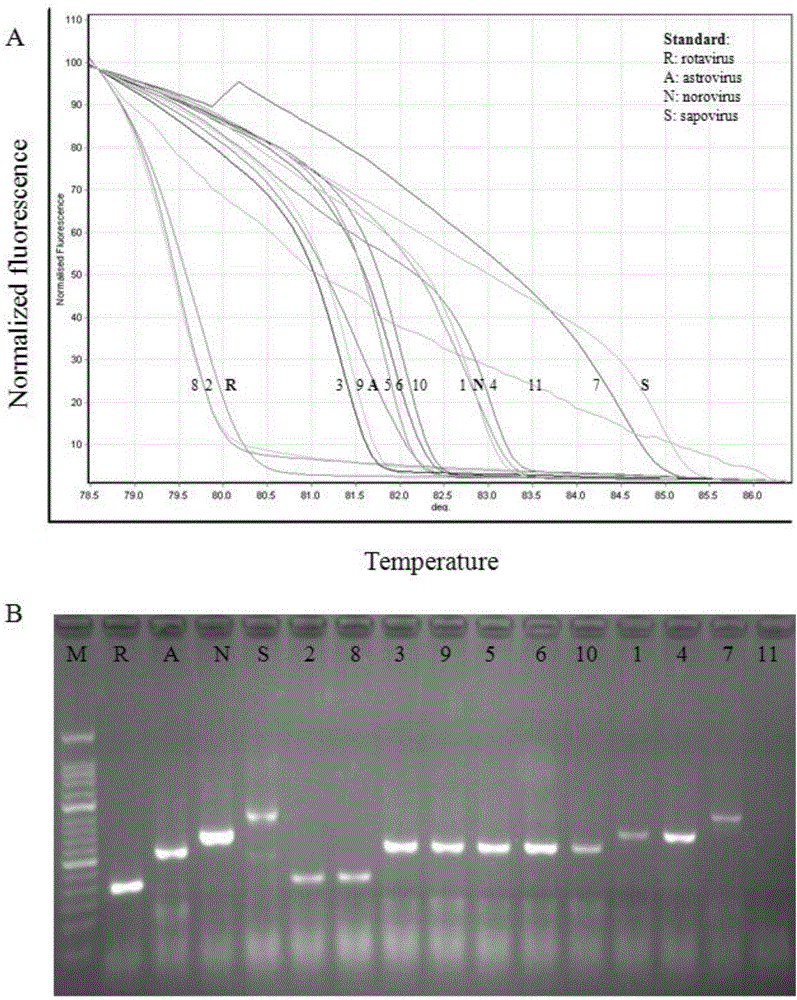
[0064] Example 3: Sensitivity analysis of the kit for detecting four diarrhea viruses based on HRMA technology
[0065] 1. Method:
[0066] (1) Sample processing: Use the above-mentioned viral RNA transcribed in vitro as a template for detection, use a micro-ultraviolet spectrophotometer to measure the concentration of the purified RNA, calculate the copy number of the initial RNA template by molecular weight, and then make it 10 times in turn Gradiently diluted to single copy number, a total of 9 gradients, the copy number is: 1×10 8 to 1×10 0 .
[0067] (2) Parallel control experiment: respectively select the HRMA detection kit of the present invention and common RT-PCR technology (basic reagents are purchased from Treasure Bioengineering (Dalian) Co., Ltd., article number DRR055A) to the above-mentioned 9 gradient dilution samples and 1 Negative controls were used for amplification testing.
[0068] HRMA detection: The detection method described in Example 2 was used to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com