Method for detecting FOXH1 gene SNP locus rs750472 genotype
A genotype and locus technology, applied in the field of detection of the FOXH1 gene SNP locus rs750472 genotype, can solve the problems of heavy economic burden on children's families, increase in abnormal cardiac function, etc., and achieve rapid detection, high sensitivity, and good specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Preparation of Example 1FOXH1rs750472 Standard
[0066] To establish an HRM analysis method, the external standard required by the method must first be prepared. The standard should contain highly conserved and specific sequences, and high specificity of the reaction must be ensured. In this study, the wild-type and homozygous mutant DNA sequences containing the FOXH1rs750472 (c.-314T>G) site were synthesized and connected to the plasmid vector pUC57 by gene recombination technology to construct the wild-type and pure mutants of the recombinant plasmid pUC57-rs750472. The mutants were identified by PCR and sequencing, and finally quantified as the standard for the method to be established, laying the foundation for the next method and evaluation.
[0067] 1. Construction and transformation of recombinant negative and positive plasmid pUC57-rs750472
[0068] 1. Synthesize FOXH1rs750472 (c.-314T>G) wild-type and homozygous mutant DNA sequences. The sequence synthesized i...
Embodiment 2
[0106] Example 2 Establishment of HRM-PCR detection method for FOXH1rs750472
[0107] 1. Preparation of samples to be tested
[0108] Genomic DNA from peripheral blood was extracted from 30 samples and used as a template for PCR amplification of FOXH1 gene.
[0109] (1) Take 1ml whole blood, add 3ml TE, invert and mix well, let stand for 10min, centrifuge at 8000rpm for 5min, discard the supernatant;
[0110] (2) Repeat the above steps 2-3 times until the precipitate is white;
[0111] (3) Add 900 μl of 10% SDS and 10 μl of 10 mg / ml proteinase K, and bathe in water at 55°C for 1 hour;
[0112] (4) Cool the centrifuge tube to room temperature, add an equal volume of phenol / chloroform / isoamyl alcohol mixture (the volume ratio of phenol, chloroform and isoamyl alcohol is 25:24:1), mix well, and centrifuge at 12000rpm for 10min;
[0113] (5) After carefully aspirating the supernatant, add an equal volume of phenol / chloroform / isoamyl alcohol mixture (the volume ratio of phenol, ...
Embodiment 3
[0170] Example 3 Application of detection method of the present invention to detect sample gene mutation
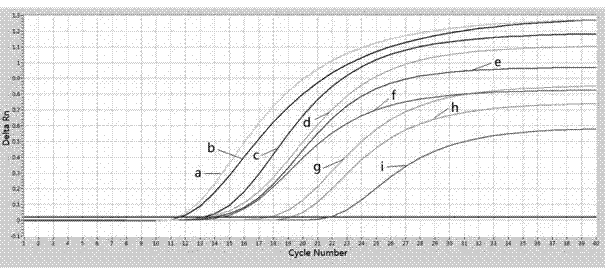
[0171] Using the optimized detection conditions in Step 4 of Example 2, HRM analysis was performed on 30 samples, compared with the Tm value of the positive plasmid, and the mutation type in the samples was determined accordingly. After the HRM analysis, agarose gel electrophoresis was performed, and one sample was randomly selected for each genotype according to the HRM analysis results for Sanger sequencing verification at Sangon Biotech (Shanghai).
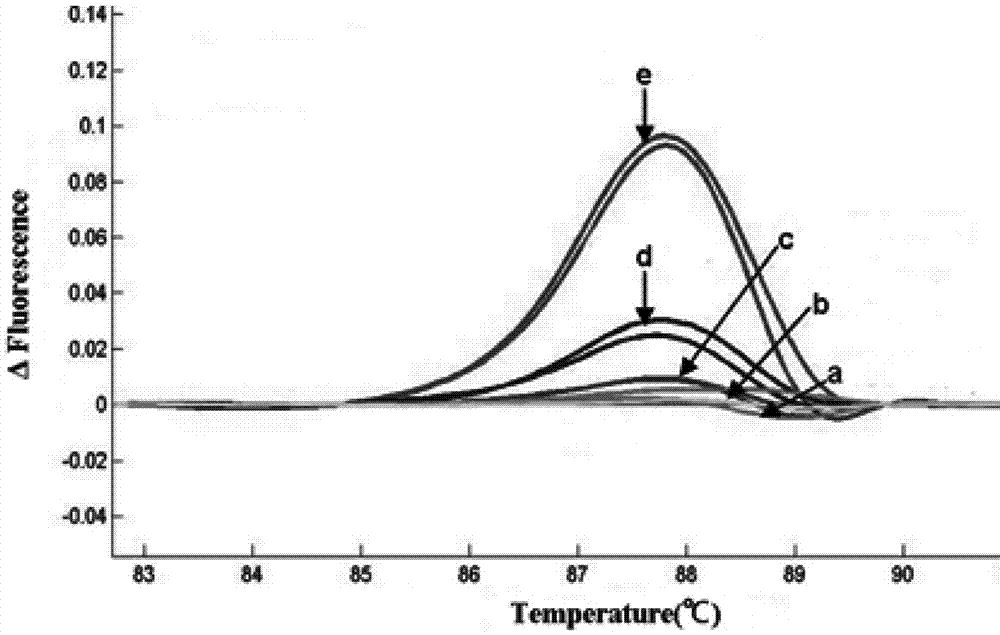
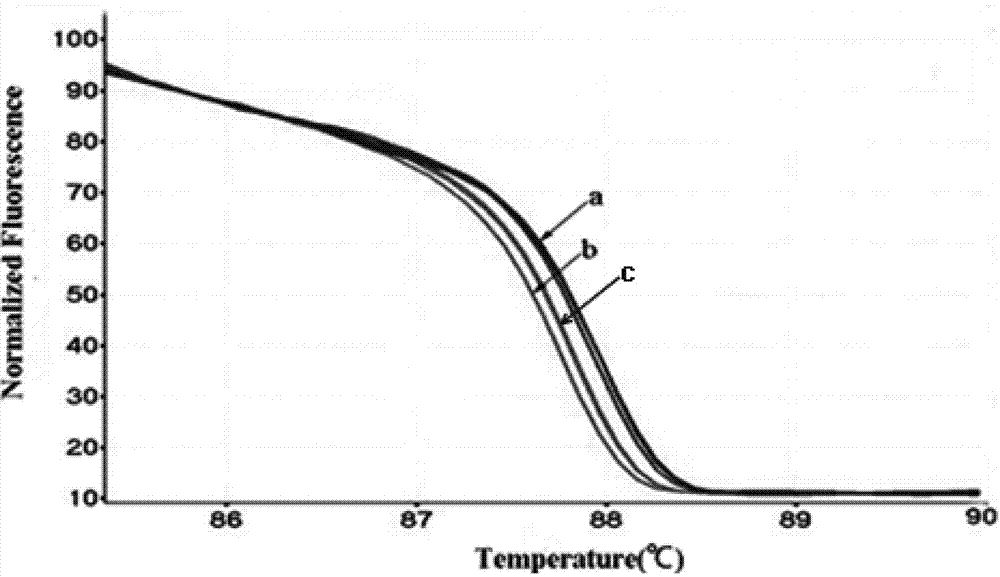
[0172] The results of HRM analysis are shown in Table 3 and image 3 , the data showed that among the 30 samples, 5 cases of FOXH1 gene rs750472 were T>C heterozygous mutant, 3 cases were T>C homozygous mutant, and the remaining 22 cases were wild type.
[0173] For Sanger sequencing results, see Figure 4 ; Wherein, a is the sequencing result of sample number 4, b is the sequencing result of sample number 16, and c is the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com