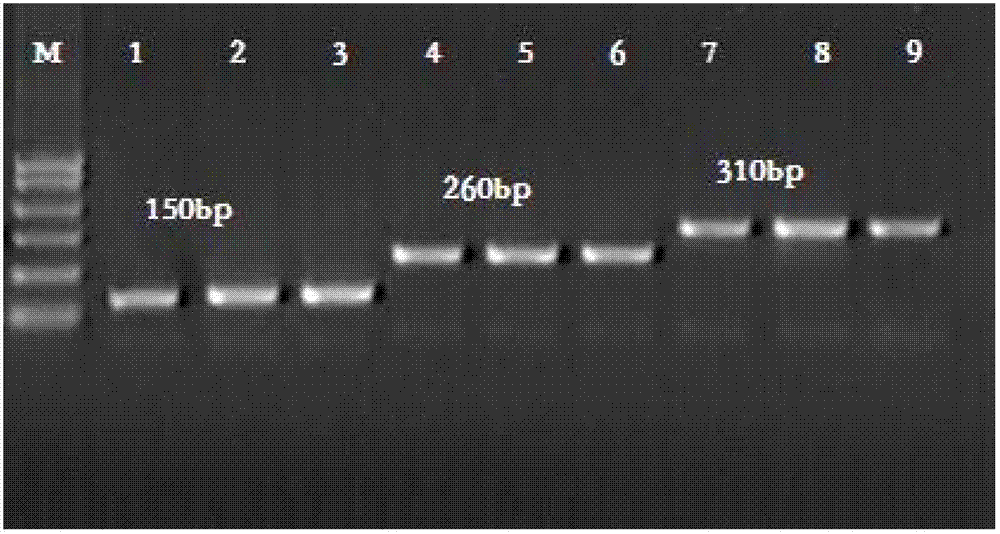
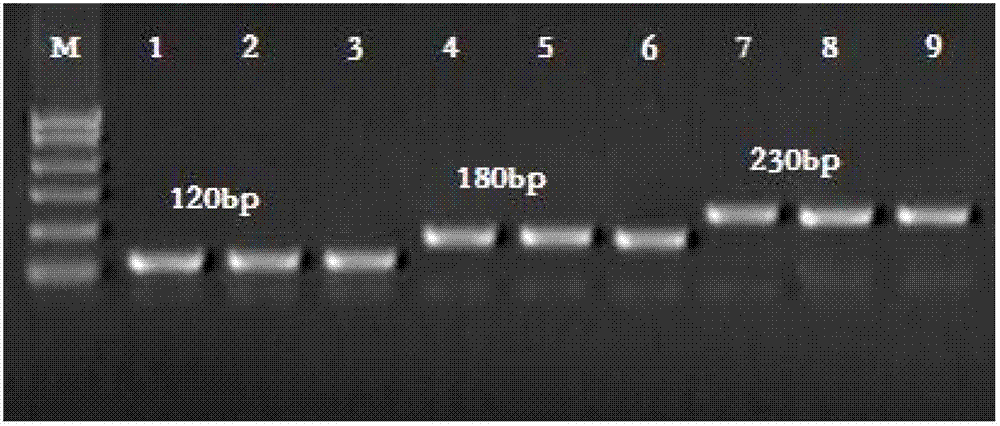
Kit and method for detecting mutant alpha-Mediterranean anemia genes through HRM (high resolution melting) method
A thalassemia and mutant technology, applied in the direction of fluorescence/phosphorescence, material excitation analysis, etc., can solve the problems of cumbersome test operations, expensive instruments and reagents, high cost, etc., to overcome hybridization or sequencing operations, complexity, short time effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] 1. Preparation of whole blood sample target nucleic acid & negative control DNA
[0076] EDTA or sodium citrate anticoagulated whole blood collected clinically from 6 cases (including a normal sample as a negative control sample), and gently invert the glass tube for 5 to 10 times to mix.
[0077] Take a 1.5ml clean centrifuge tube and mark the tube cap with a Mark pen. Add 300 μl of anticoagulated whole blood into a centrifuge tube, add 1ml of sterilized double distilled water, mix well, and place at room temperature for 3 minutes.
[0078] Centrifuge at 3000 rpm for 5 minutes at room temperature. Discard the supernatant and keep the maroon precipitate.
[0079] Repeat the above steps 1 time. Remove as much residual liquid as possible.
[0080] Add DNA extraction solution (5% Chelex-100, 0.25ug / mL proteinase K) and mix well. Water bath at 100°C for 10 minutes, centrifuge at 10,000 rpm for 5 minutes, and store at -20°C for later use. DNA extracted from normal samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com