Method and kit for detecting human PIK3CA gene mutation
A kit and technology for detecting people, applied in the fields of biotechnology and medicine, to achieve the effect of convenient and fast detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: Selection of samples and extraction of genomic DNA
[0062] The collected genomic DNA comes from whole blood, cells, fresh tissue, and paraffin-fixed tissue. The extraction method refers to the operation manual provided by the kit, and some optimizations have been made at the same time.
[0063] Whole blood genomic DNA of healthy people was used as the wild-type control of the experiment, and was extracted using the BloodGen Mini Kit of Kangwei Century;
[0064] The genomic DNA derived from the OMC-1 cell line was used as a control for the homozygous mutant type (1633G>A) of exon 9 of the PIK3CA gene in the experiment,
[0065] The genomic DNA derived from the MCF-7 cell line was used as the control of the PIK3CA gene codon No. 9 heterozygous mutant (1633G>A) in the experiment, and was extracted using the DNA FlexGen DNAKit of Kangwei Century;
[0066] The genomic DNA derived from the SK-OV-3 cell line was used as a control for the homozygous mutant type (31...
Embodiment 2
[0076] Example 2: Identification of mutation sites
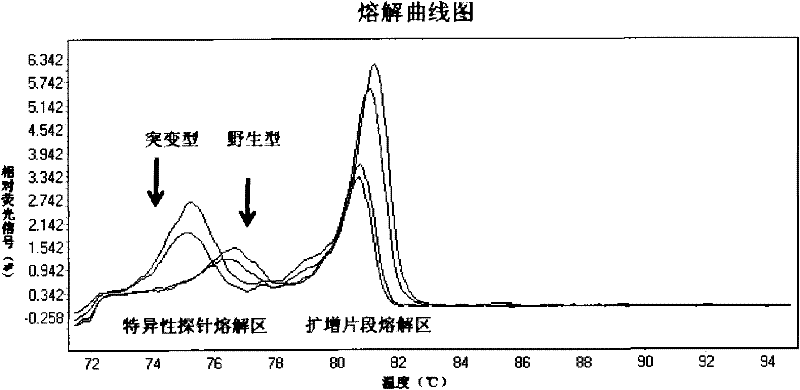
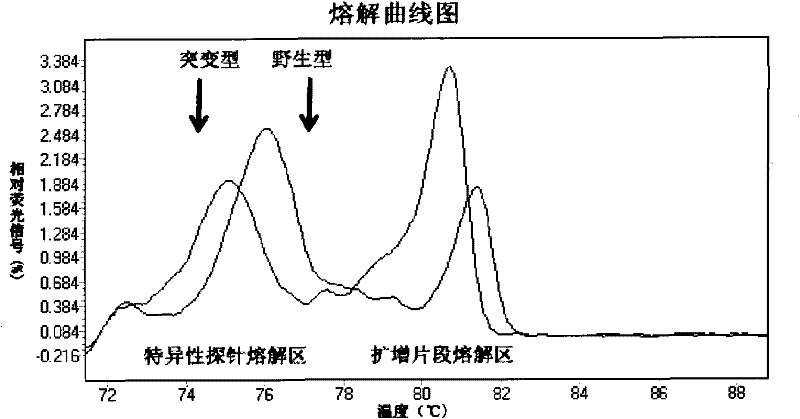
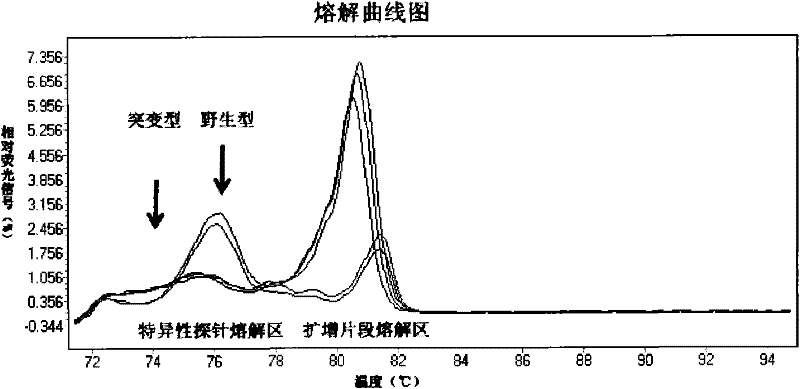
[0077] HRM technology was used to detect the genotypes of the E542K and E545K codons of exon 9 of the PIK3CA gene and the H1047R codon of exon 20 of the PIK3CA gene.
[0078] 1. The specific primers and probes for exon 9 are as follows:
[0079] SEQ ID No: 5, Forward primer:
[0080] 5'-CAGCTCAAAGCAATTTCTACACGAGAT-3'
[0081] SEQ ID No: 6, Reverse primer: 5'-TTAGCACTTACCTGTGACTCCATAGA-3'
[0082] SEQ ID No: 3, specific probe: 5'-TCTTTCTCCTGCTCAGTGAT-3'
[0083] The specific primers and probes for exon 20 are as follows:
[0084] SEQ ID No: 11, Forward primer: 5'-GCAAGAGGCTTTGGAGTATTTCAT-3'
[0085] SEQ ID No: 12, Reverse primer: 5'-TGCTGTTTAATTGTGTGGAAGATCC-3'
[0086] SEQ ID No: 4, specific probe: 5'-CCACCATGATGTGCATCATTC-3'
[0087] 2. Amplify the fragments near the E542K and E545K codons of exon 9 and the H1047R codon of exon 20 respectively by PCR; prepare the mixed solution: add 1 μl of the genomic DNA solution p...
Embodiment 3
[0091] Example 3: Kit Sensitivity Verification
[0092] Taking exon 9 of the PIK3CA gene as an example, the homozygous mutant OMC-1 genome and the wild-type genome extracted from whole blood were mixed in a certain proportion, so that the homozygous mutant genome accounted for 0.05% of the total DNA amount. The concentration of the above mixed genomic DNA and wild-type genomic DNA was adjusted to 10 ng per microliter.
[0093] 1. The specific primers and probes for exon 9 are as follows:
[0094] SEQ ID No: 5, Forward primer:
[0095] 5'-CAGCTCAAAGCAATTTCTACACGAGAT-3'
[0096] SEQ ID No: 6, Reverse primer: 5'-TTAGCACTTACCTGTGACTCCATAGA-3'
[0097] SEQ ID No: 3, specific probe: 5'-TCTTTCTCCTGCTCAGTGAT-3'
[0098] 2. Amplify some fragments near the E542K and E545K codons of exon 9 by PCR; prepare the mixed solution: add 1 μl of the genomic DNA solution prepared before, 2 μl of PCR buffer (10×), 1.6 μl of dNTP, 0.1 μl of Taq DNA polymerization Enzyme, 0.4 μl of forward prime...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com