Method for detecting lncRNA (long noncoding RNA) in plasma
A medium-to-long-chain, non-coding technology, which is applied in the field of detection of long-chain non-coding in plasma, which can solve the problems of easy degradation of RNA, no extraction of lncRNA, low content of total RNA, etc., and achieve the effect of high content extraction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] A method for detecting lncRNA in plasma, the steps are as follows:
[0025] 1. After anticoagulation of whole blood with EDTA, centrifuge at 3000 rpm for 10 minutes at room temperature, take 300 μl of the upper plasma and transfer it to the first RNase-free centrifuge tube, add 900 μl of Trizol LS reagent, vortex for 10 seconds, and stand at room temperature for 5 minutes; at 4°C, Centrifuge at 12000rpm for 10 minutes. The sample is divided into two layers. The upper layer is a pink homogenized liquid, and the lower layer is a darker viscous impurity layer. Transfer the upper layer (about 1000 μl) to a second RNase-free centrifuge tube and discard the precipitate. , to remove impurities such as polysaccharides and mucins;
[0026] 2. Add 0.2ml of chloroform to the second RNase-free centrifuge tube, vortex for 10 seconds, let stand at room temperature for 5 minutes, then centrifuge at 12,000rpm for 15 minutes at 4°C, and absorb the upper aqueous phase (prefer less than e...
Embodiment 2
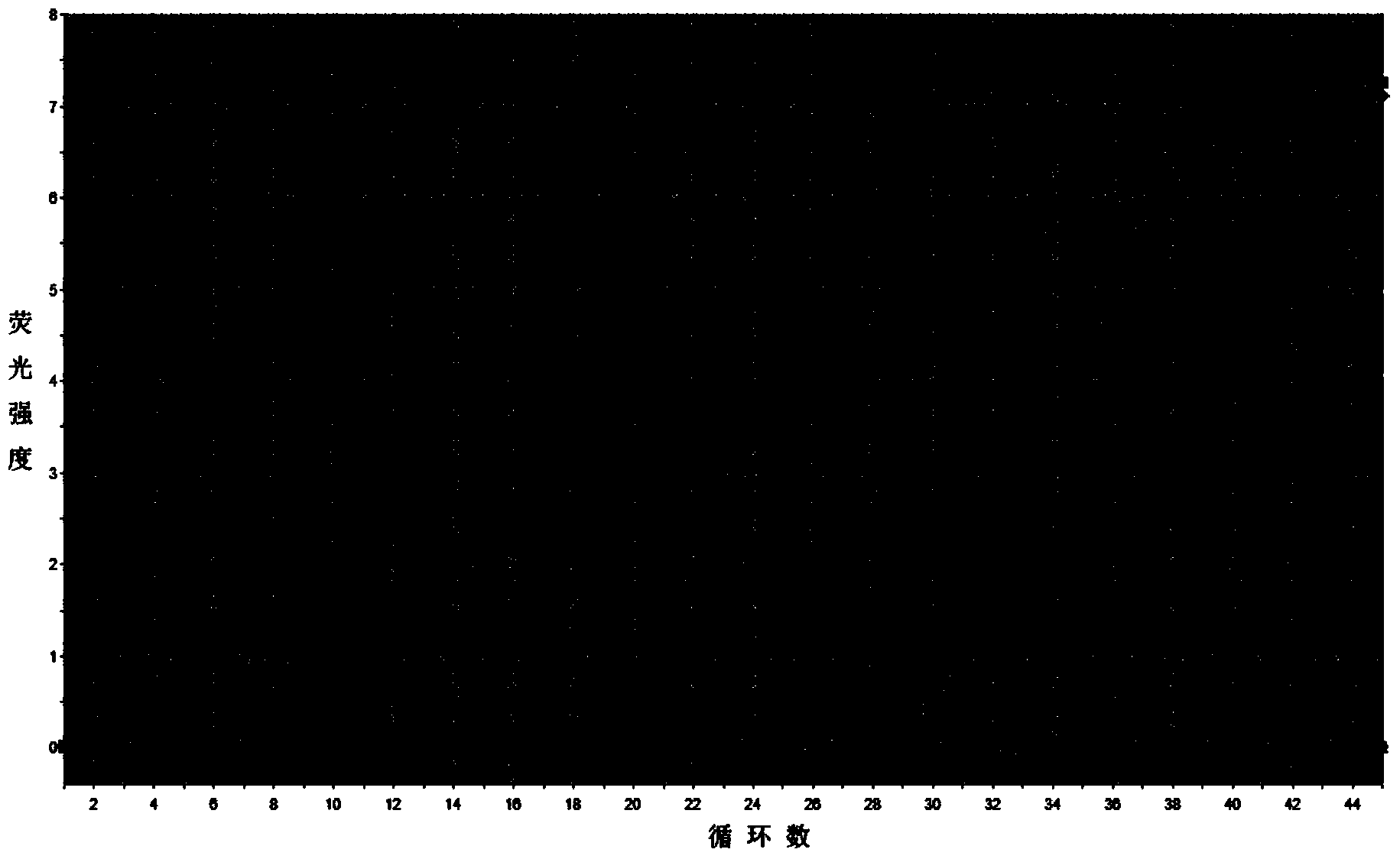
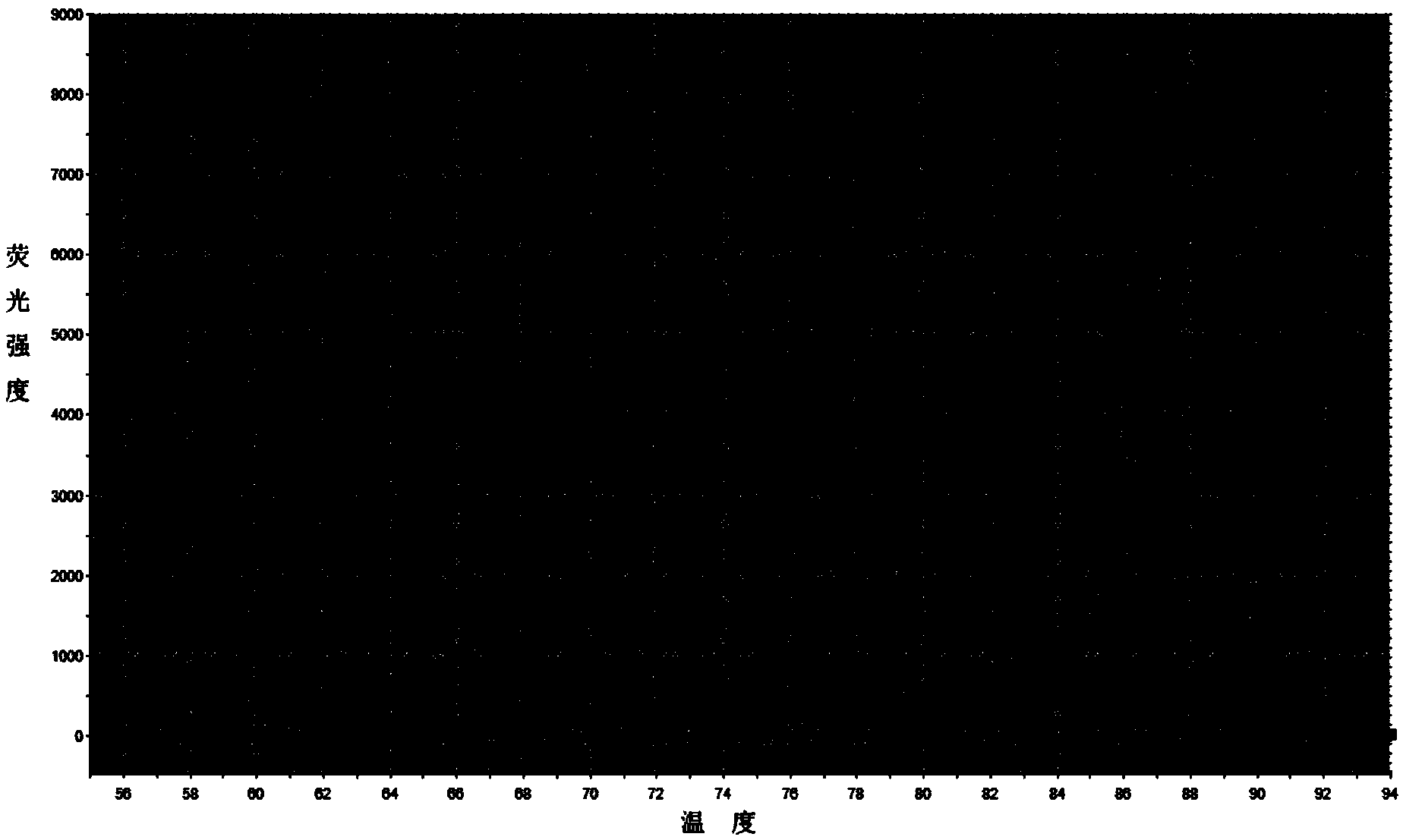
[0032] The method is basically the same as in Example 1, except that in step 9, the upstream and downstream primers for amplification are replaced by AA174084-specific amplification primers for RMRP-specific amplification primers, and the amplification curve and solution of the expression level of AA174084 in plasma are detected. From the amplification curve, it can be obtained that the Ct values of the two plasma samples detected are 34.21 and 34.91 respectively; from the melting curve, it can be known that the curve is a narrow single peak, and it can be seen that the Ct values of each plasma sample AA174084 was amplified specifically without the interference of primer dimers and heterogeneous bands.
Embodiment 3
[0034] The method is basically the same as in Example 1, except that in step 9, the upstream and downstream primers for amplification are replaced by BM709340-specific amplification primers for RMRP-specific amplification primers, and the amplification curve and solution of the expression level of BM709340 in plasma are detected. From the amplification curve, it can be obtained that the Ct values of the two plasma samples detected are 27.87 and 30.46 respectively; from the melting curve, it can be known that the curve is a narrow single peak, and it can be seen that the Ct values of each plasma sample are BM709340 was amplified specifically without the interference of primer dimers and heterogeneous bands.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com