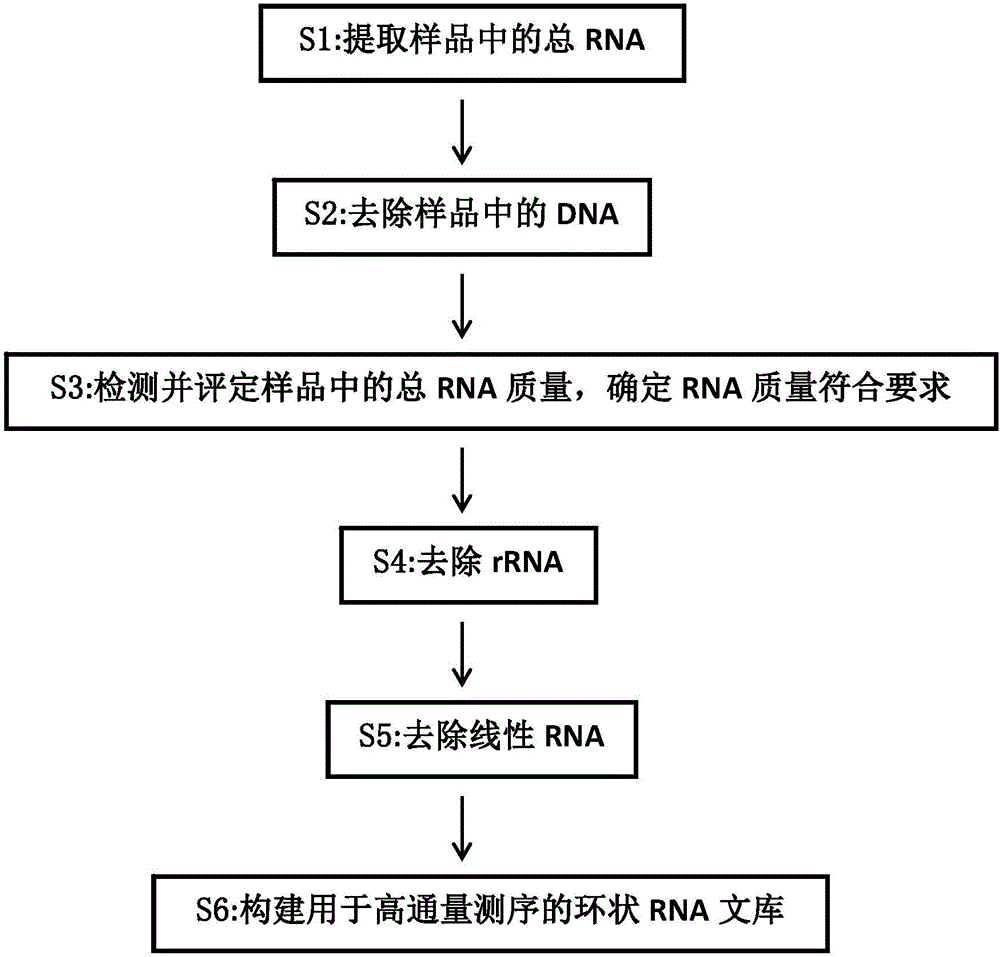
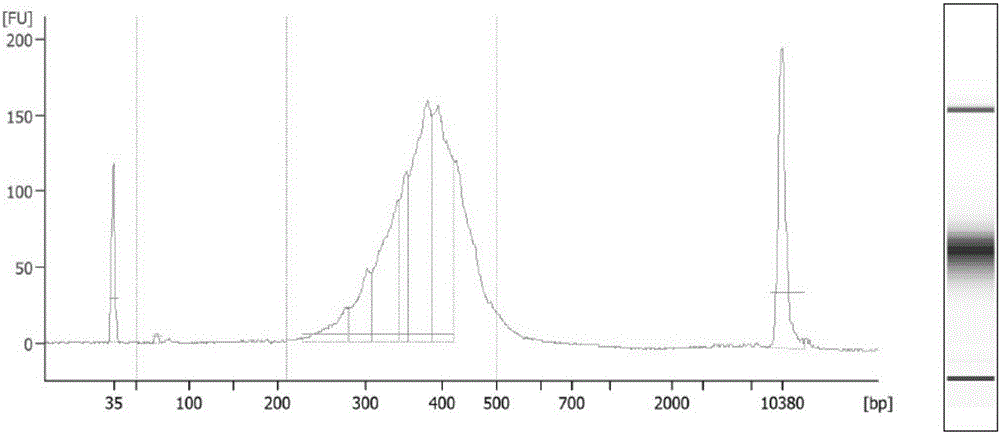
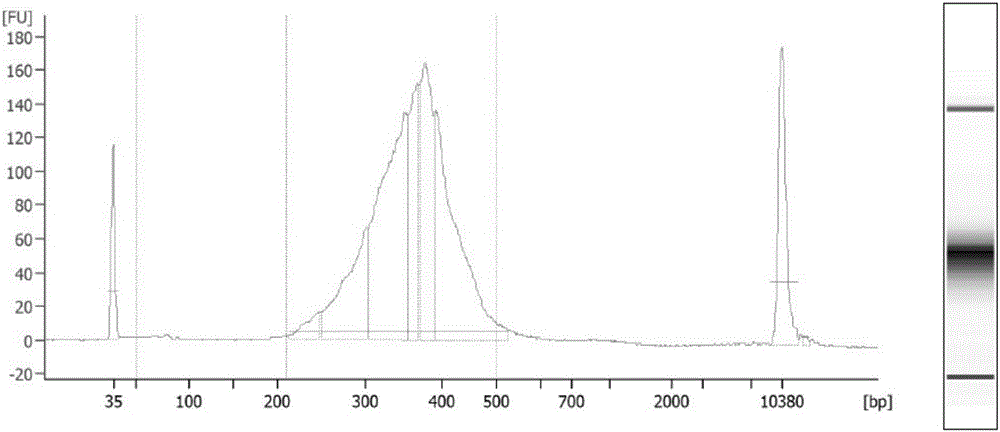
Establishing method of circular RNA high-throughput sequencing library and kit thereof
A construction method and technology for sequencing libraries, which are applied in the field of circular RNA high-throughput sequencing library construction methods and kits, can solve the problems of high price and limited circular RNA research progress, and achieve low-cost, circular RNA The construction method of high-throughput sequencing library is efficient and the effect of low residue ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118] Example 1 Construction of circular RNA high-throughput sequencing library in cell samples
[0119] (S1) Extraction of total RNA in cell samples
[0120] Take cell sample 1×10 7 Wash once with PBS at 4°C, then add 1mL Trizol to each well of the 6-well plate, and repeatedly pipette 10 times with a 1mL pipette tip. Collect the samples into EP tubes, add 200 μL of chloroform, mix up and down for 30 seconds and let stand for 3 minutes. Then centrifuge at 4° C. and 12,000 rpm for 15 minutes, and the solution lysate is divided into three layers, wherein the upper layer is RNA dissolved in the water phase. Take the upper layer solution and add an equal volume of isopropanol to it, mix well and let stand at 4°C for 10 min. After that, centrifuge again at 4° C. and 12000 rpm for 10 min and remove the supernatant. Then add 1 mL of 75% ethanol to the pellet and mix up and down to resuspend the pellet. Then centrifuge at 4° C. and 12000 rpm for 10 min, remove the supernatant, a...
Embodiment 2
[0172] Example 2 Construction of circular RNA high-throughput sequencing library in fresh liver cancer tissue samples
[0173] (S1) Extraction of total RNA in fresh liver cancer tissue samples
[0174] Take 50 g of fresh liver cancer tissue samples, wash once with PBS at 4°C, and cut the tissue into pieces. Then add 1mL Trizol and homogenize 15 times with a hand-held high-speed disperser. Afterwards, the samples were collected into EP tubes and 200 μL of chloroform was added, mixed up and down for 30 seconds, and then allowed to stand for 3 minutes. Then centrifuge at 4° C. and 12,000 rpm for 15 minutes, and the solution lysate is divided into three layers, wherein the upper layer is RNA dissolved in the water phase. Take the upper layer solution and add an equal volume of isopropanol to it, mix well and let stand for 10min. After that, centrifuge again at 4° C. and 12000 rpm for 10 min and remove the supernatant. Then add 1 mL of 75% ethanol to the pellet and mix up and d...
Embodiment 3
[0225] Example 3 Construction of circular RNA high-throughput sequencing library in FFPE samples
[0226] (S1) Extraction of total RNA in FFPE samples
[0227] Slice the FFPE sample into 8 μm thick slices, and immediately transfer 7 slices to a 1.5mL centrifuge tube. Then 1 mL of xylene was added and vortexed vigorously for 30 s, and centrifuged at 14000 rpm for 2 min. Then add 1 mL of ethanol to the sample and vortex for 30 s, then centrifuge at 14,000 rpm for 2 min, remove the supernatant, and keep the precipitate. Dry at 37 °C for 15 min to remove ethanol. Then add 200 μL lysate and 20 μl Proteinase K to the pellet and vortex to mix. This was followed by a water bath at a temperature of 55°C for 15 min, followed by a water bath at a temperature of 80°C for 15 min. After brief centrifugation at low speed, add 200 μL of buffer to the sample and vortex for 20 seconds. Then add 600 μL absolute ethanol to the sample and vortex mix for 20 s. Then transfer the mixture to an ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com