A method for developing Ainaxiang ssr primers based on transcriptome sequencing
A technology of Ainaxiang and primer pairs, which is applied in the fields of molecular biology and bioinformatics, and can solve the problems of insufficient yield, low content of medicinal ingredients, and a small number of Ainaxiang SSR primers, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 RNA-seq analysis and design of SSR primers
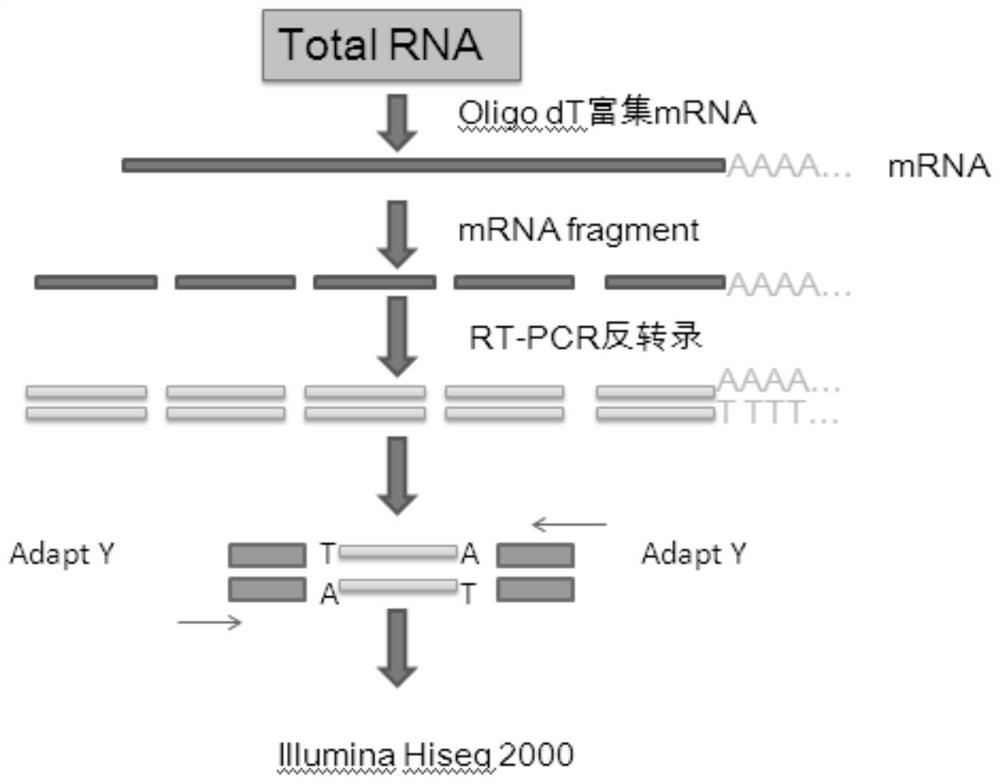
[0043] 1. RNA-seq analysis sample processing and sequencing process see figure 1 . The specific method is as follows:
[0044] For the extraction of total RNA of Ainaxiang, eukaryotes build a library with an initial amount of 5ug total RNA;
[0045] After the mRNA was separated by the magnetic bead method, the ion interrupted the mRNA (Truseq TM RNA sample prep Kit);
[0046] Recover 200-700bp fragments, reverse transcribe and synthesize double-stranded cDNA, fill in, add A to the 3' end, and connect the index adapter (Truseq TM RNA sample prep Kit);
[0047] Library enrichment, PCR amplification for 15 cycles;
[0048] 2% agarose gel recovery target band (Certified Low Range Ultra Agarose);
[0049] TBS380 (Picogreen) quantified, mixed according to the data ratio on the machine;
[0050] Perform bridge PCR amplification on cBot to generate clusters;
[0051] Hiseq2000 sequencing platform for 2*100bp sequ...
Embodiment 2
[0094] Example 2 Excavation of high-throughput SSR sites of Ainaxiang
[0095] Identification of SSR loci
[0096] First install the Perl language, download est-trimmer.pl from the http: / / pgrc.Lpk-gatersleben.de / misa website, and remove the short and long sequences in the transcriptome sequence; from http: / / www.bioinformatics , download CD-HIT software from org / cd-hit / to remove redundant sequences.
[0097] Download from http:pgrc.lpk-gatersleben.de / misa website and use MISA software to identify and locate SSR in the sequence, the parameters are set as follows: single nucleotide, dinucleotide, trinucleotide, tetranucleotide, five The number of repeats of nucleotides and hexanucleotides is at least 10, 6, 5, 3, 3, 3.
[0098] Design of SSR primers
[0099] Use Primer3 to batch design SSR primers, website: http: / / sourceforge.net / projects / primer3 / files / primer3 / 1.1.4 / primer3-1.1.4-WINXP.zip / download, primer design parameters are primer length 18-22bp ,T m 55-65°C. The diffe...
Embodiment 3
[0100] Example 3 Using SSR primers to identify polymorphisms in Ainaxiang DNA from different places
[0101] Extract 9 copies of Ainaxiang DNA, use 0.8% agarose gel electrophoresis to check the quality, dilute the DNA concentration to 50ng / ul, and store it at -20°C for future use. The DNA of the materials used for primer development was used to identify the success rate of primer design by PCR. The total volume of PCR reaction is 25μl, 1.5mmol / L MgCl 2, 100 μmol / L each of the four dNTPs, 80ng each of the primers, 1.5U Taqplus DNA polymerase (high fidelity), 50ng DNA. The reaction program was: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 51-60°C for 45 s, extension at 72°C for 45 s, 32-35 cycles, and finally extension at 72°C for 5 min. After the reaction, add 2ul of sample buffer to the product, use 100bp DNA ladder as DNA molecular weight standard, use 8% non-denaturing polyacrylamide gel for electrophoresis, electrophoresis buffer level 0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com