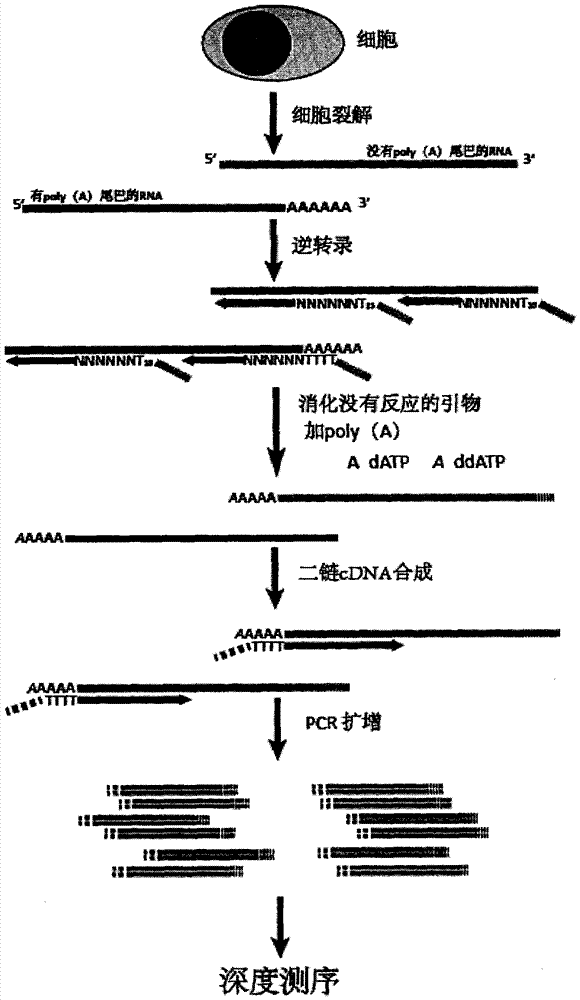
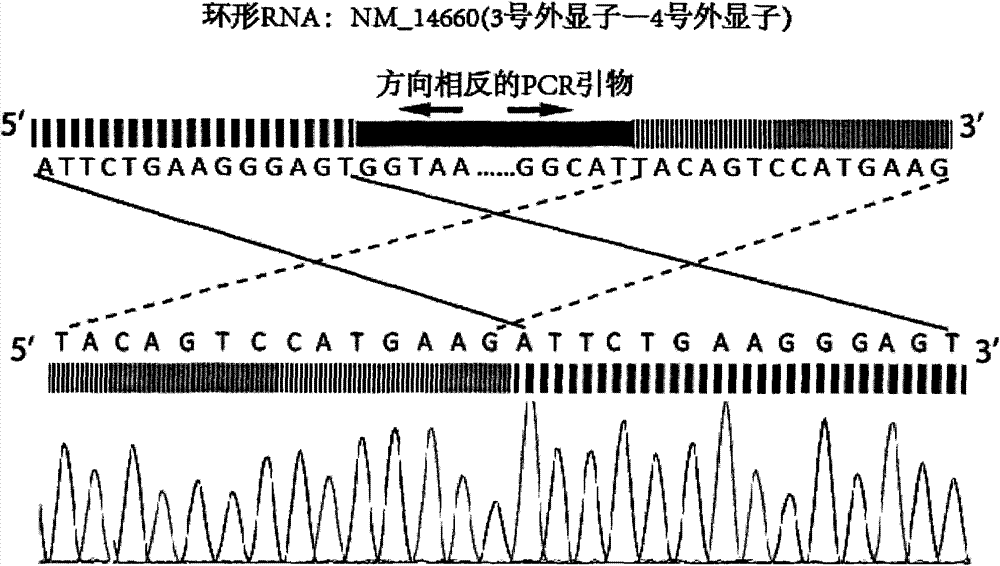
Transcriptome sequencing method
A technology of transcriptome sequencing and transcriptome, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve sequencing and other problems, achieve high technical repeatability, and eliminate technical errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] 1. Lysis of single cells:
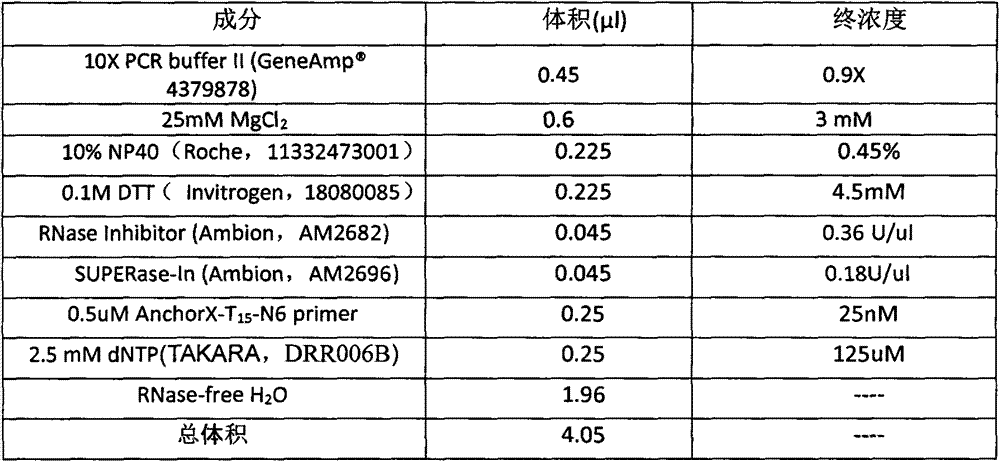
[0021] Prepare the following lysate:
[0022]
[0023] 25mM MgCl 2 It is included in the 10X PCR buffer ll package. AnchorX-T15-N6primer is a primer used in the reverse transcription process of RNA to cDNA, wherein N6 is a sequence consisting of 6 random bases, and the random bases are randomly selected from any of A, G, C, and T. T 15 It is a T homomer of 15bp. Anchor) (anchor sequence) is a synthetic 23-base length sequence that is not homologous to mammalian gene sequences.
[0024] 4.5 μl of lysate was mixed and added to the PCR tube containing single cells.
[0025] Isolation of single cells:
[0026] The cells were digested with 0.25% trypsin for 3-5 minutes, and the digestion was terminated with three times the volume of cell culture medium containing leukemia inhibitory factor (LIF). Observe and operate under a microscope in PBS buffer. Use a capillary needle with a cross-sectional diameter of 20-30 μm to aspirate single cel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com