Patents
Literature
45 results about "Pathway analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
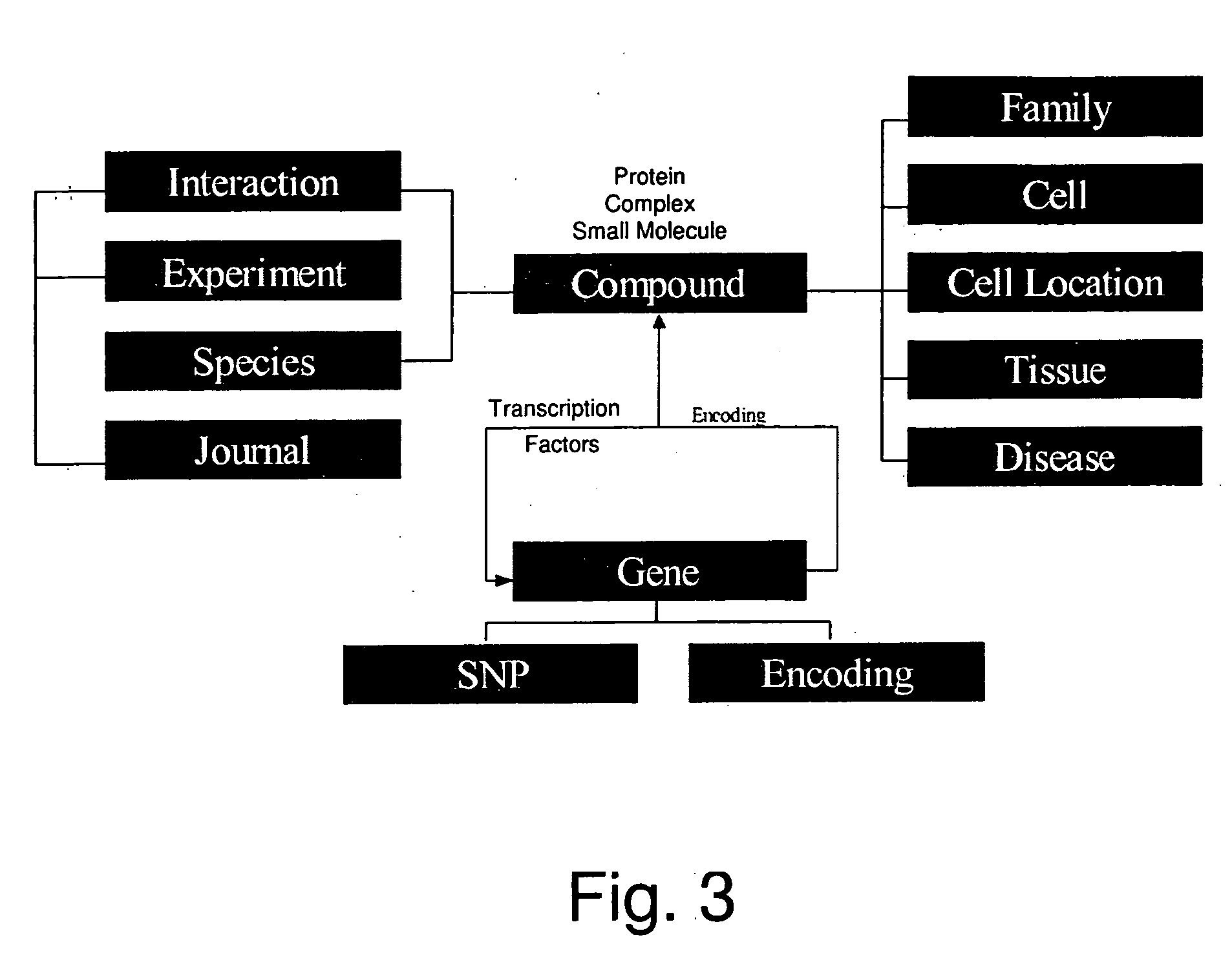
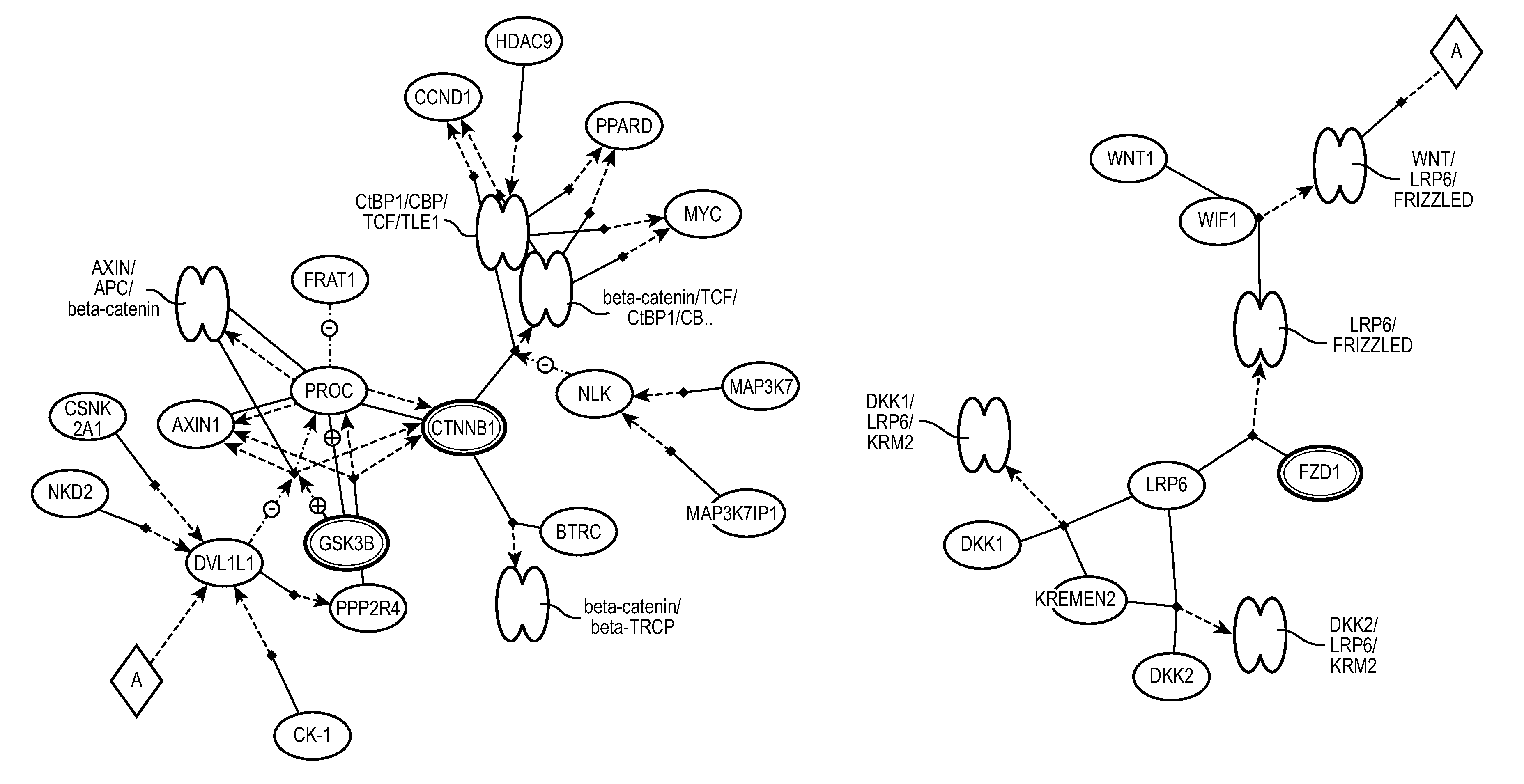
In bioinformatics research, pathway analysis software is used to identify related proteins within a pathway or building pathway de novo from the proteins of interest. This is helpful when studying differential expression of a gene in a disease or analyzing any omics dataset with a large number of proteins. By examining the changes in gene expression in a pathway, its biological causes can be explored. Pathway is the term from molecular biology which depicts an artificial simplified model of a process within a cell or tissue. A typical pathway model starts with an extracellular signaling molecule that activates a specific receptor, thus triggering a chain of protein-protein or protein-small molecule interactions. Pathway analysis helps to understand or interpret omics data from the point of view of canonical prior knowledge structured in the form of pathways diagrams. It allows finding distinct cell processes (Cellular processes), diseases or signaling pathways that are statistically associated with selection of differentially expressed genes between two samples. Often but erroneously pathway analysis is used as synonym for network analysis (functional enrichment analysis and gene set analysis).
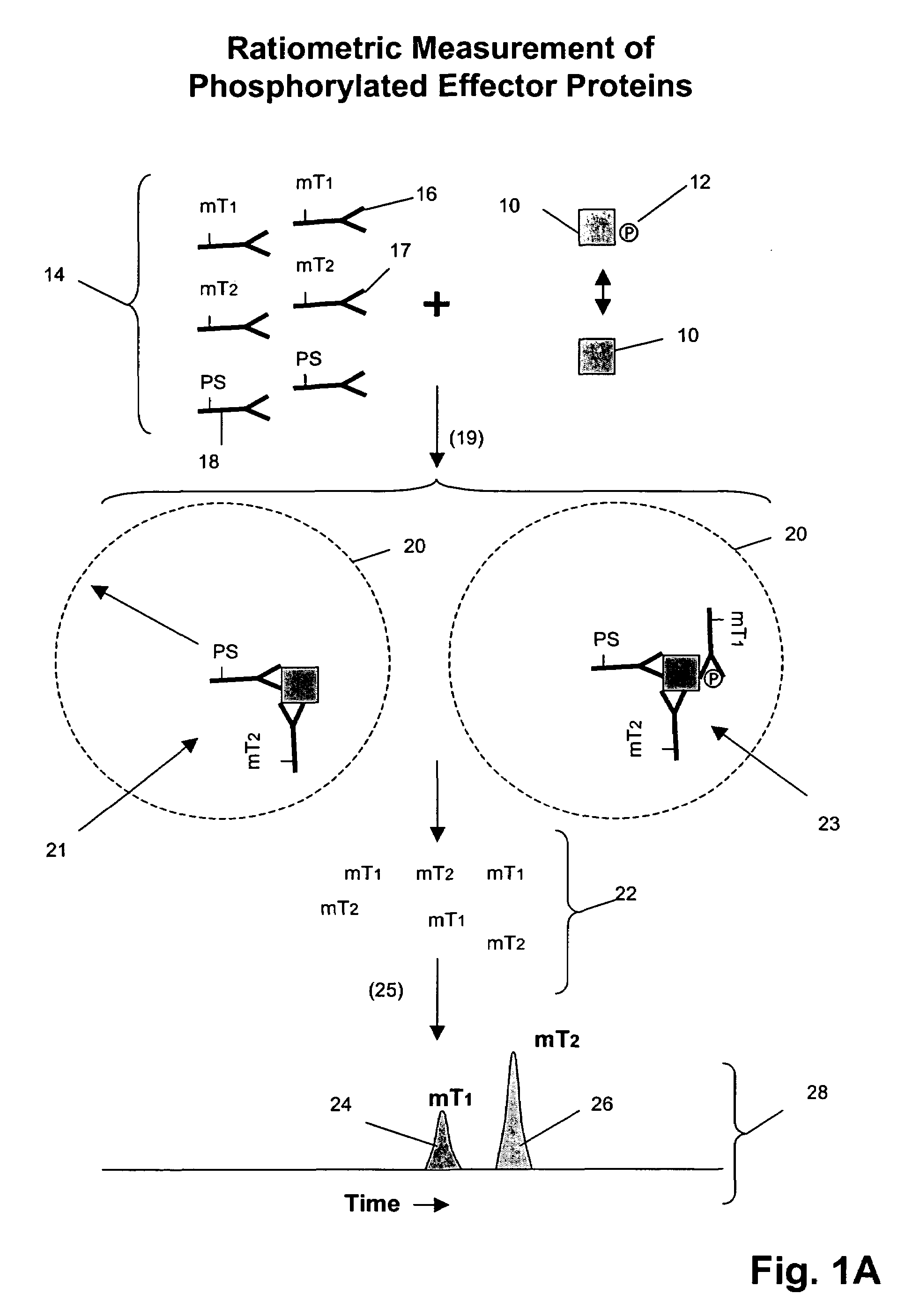
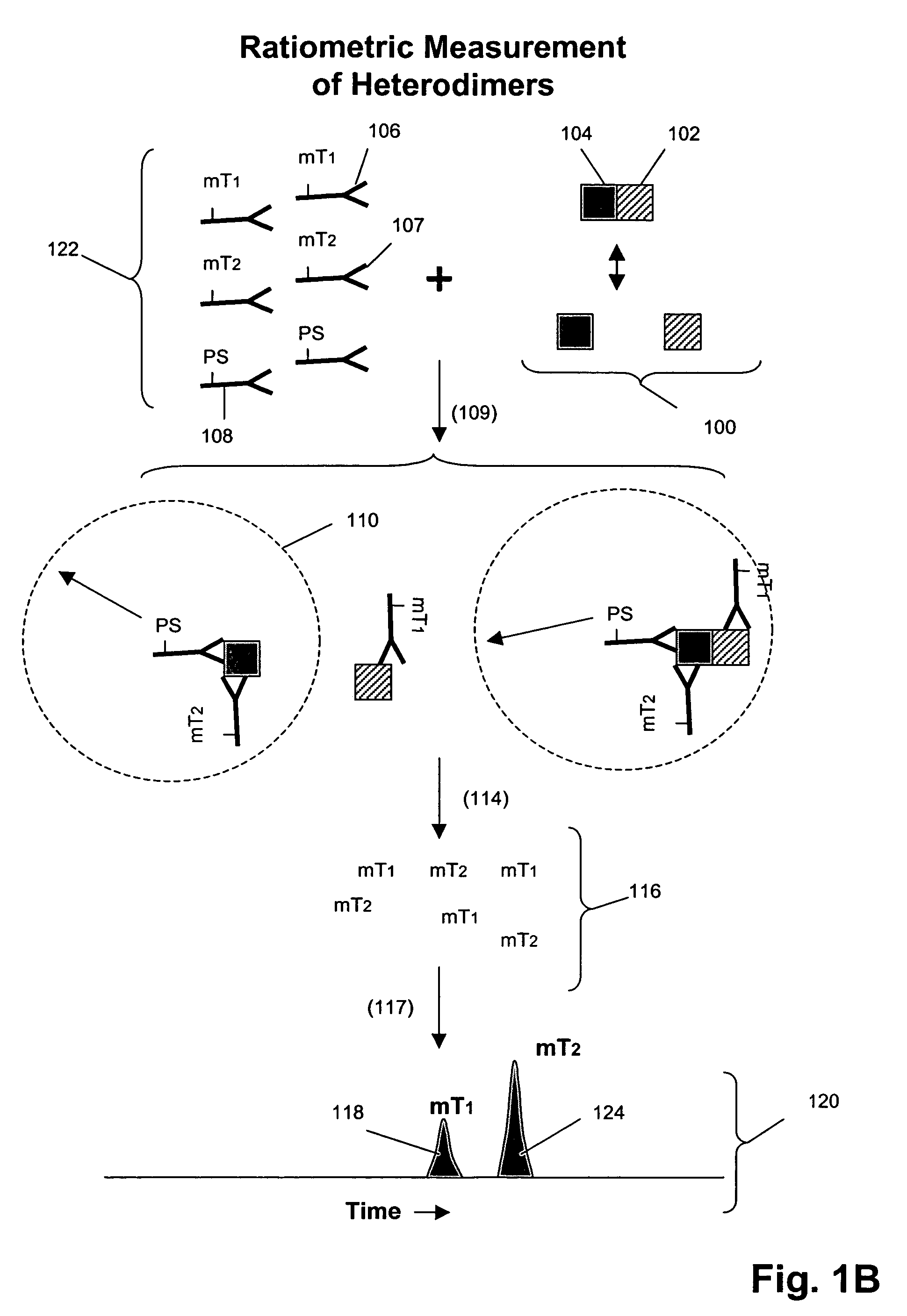
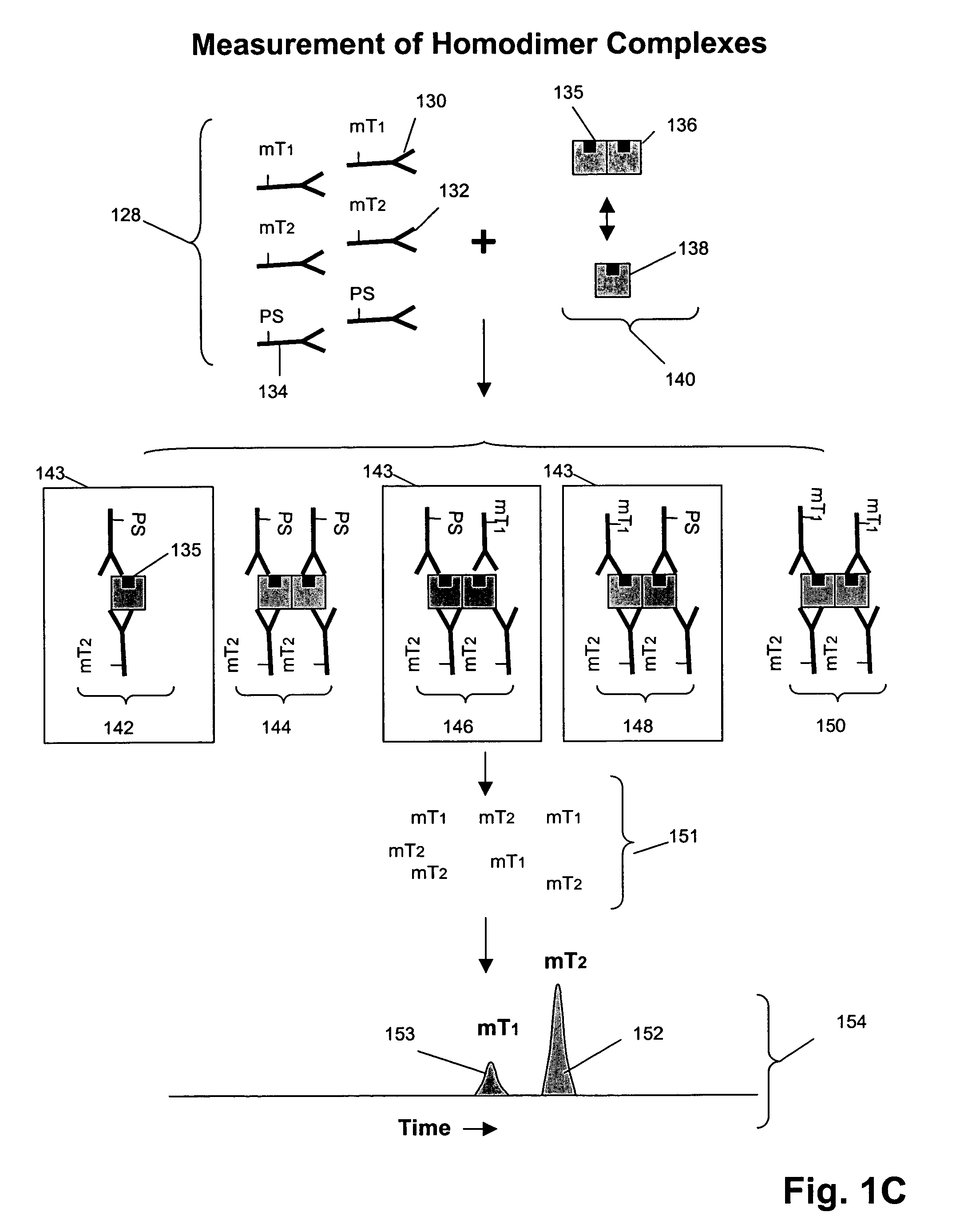
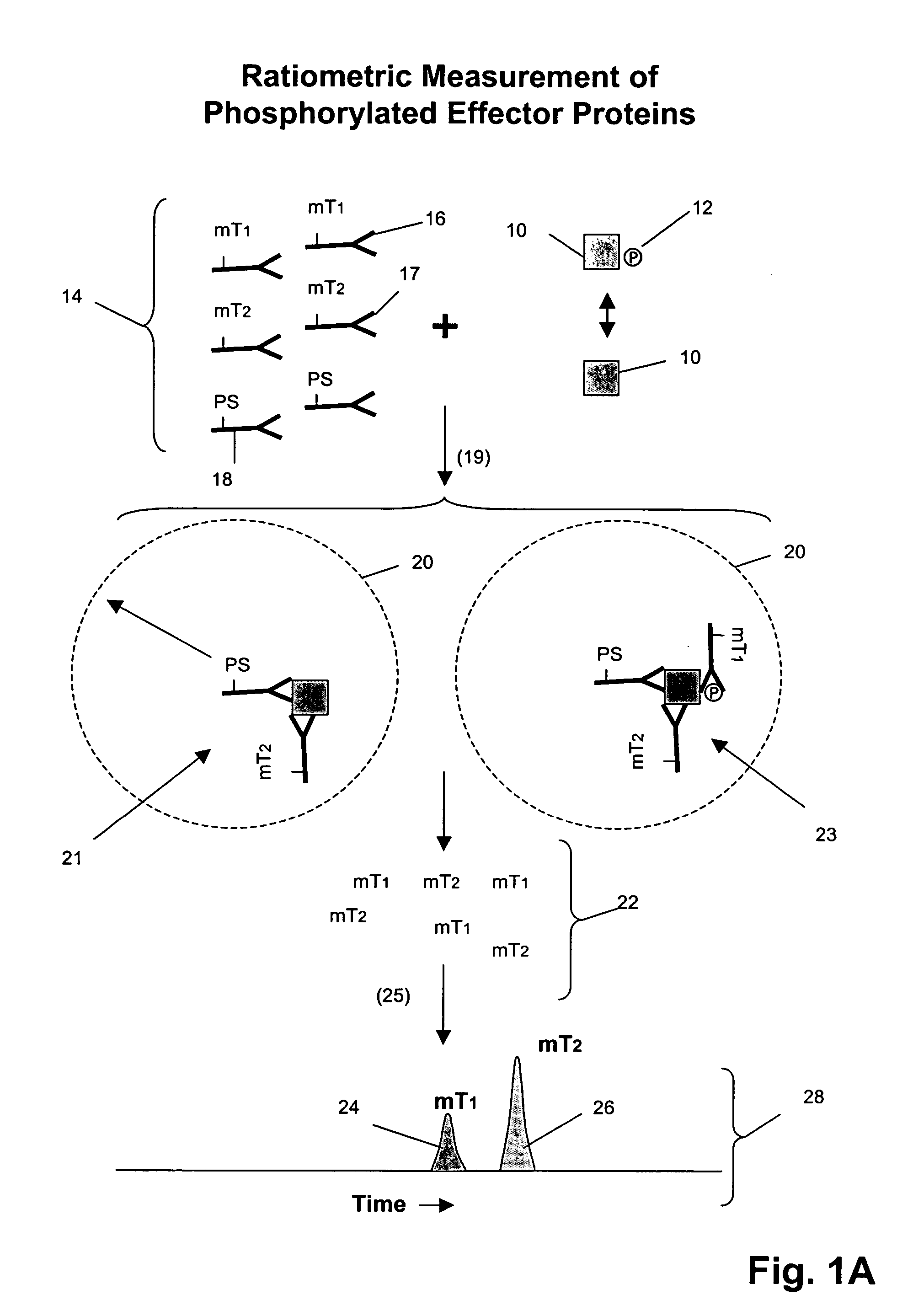
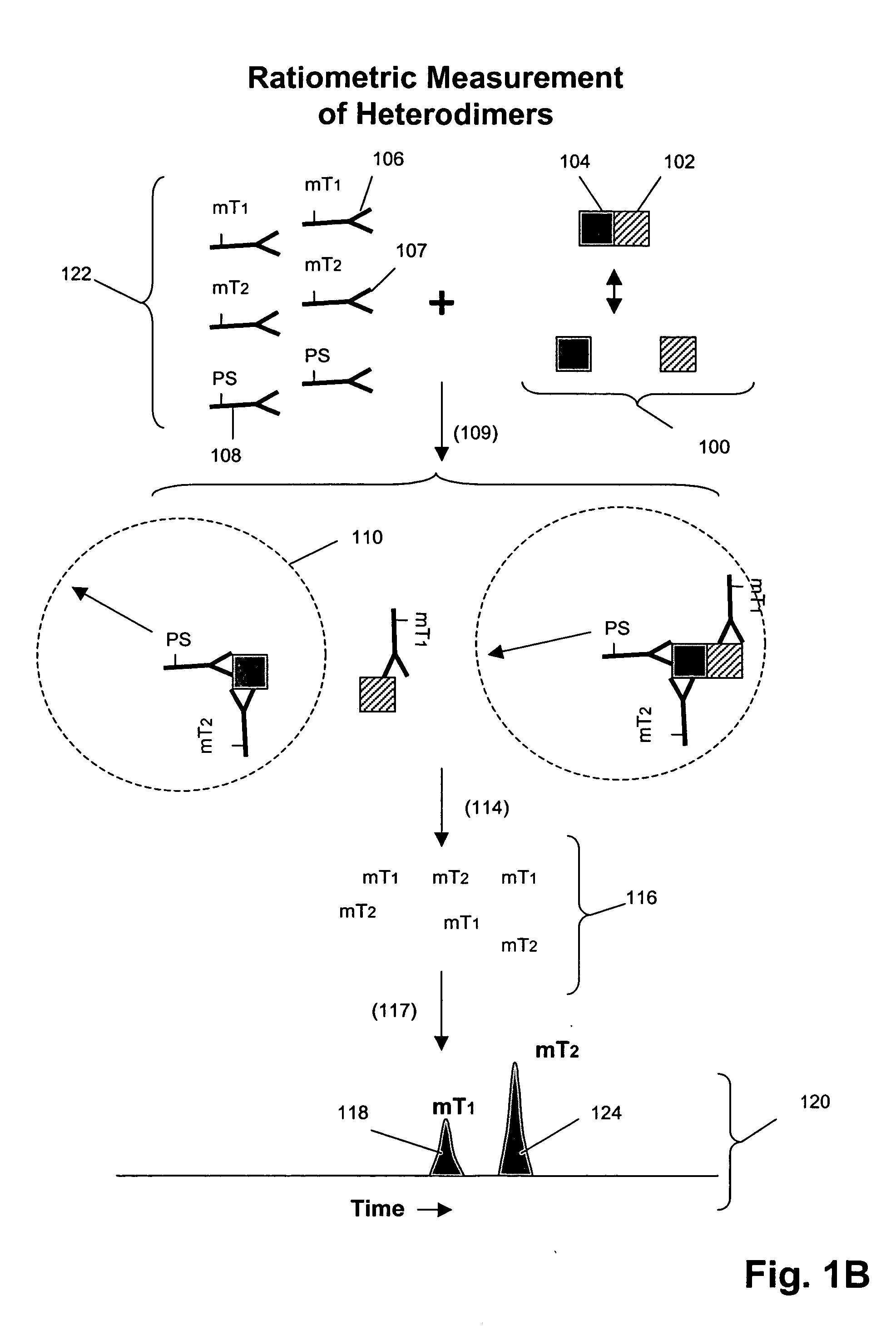
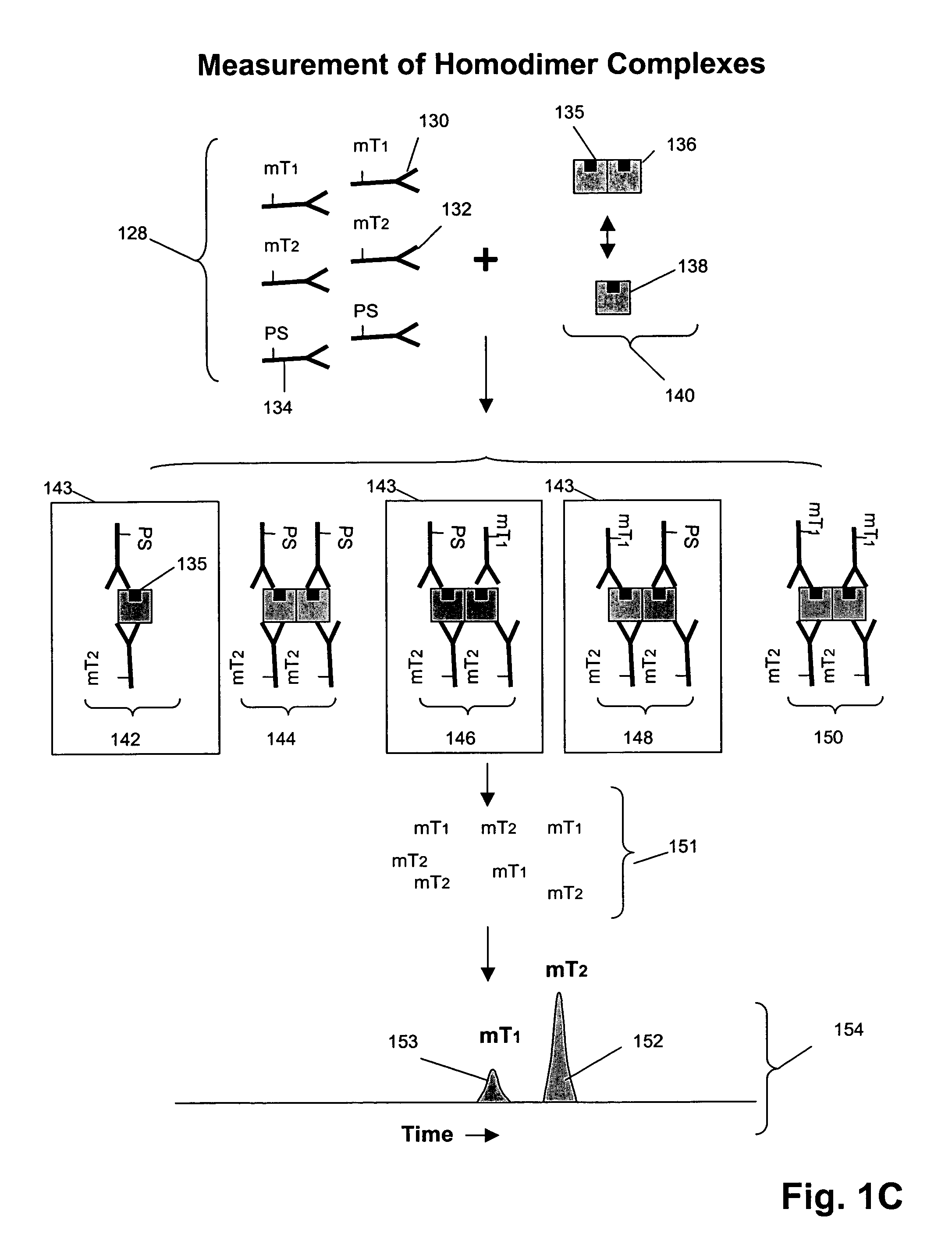
Receptor tyrosine kinase signaling pathway analysis for diagnosis and therapy
The invention provides a method for determining the activation status of receptor tyrosine kinase (RTK) pathways in either cell samples or patient samples by measuring receptor dimerization and relative amounts of protein-protein complexes or activated effector proteins that are characteristic of an RTK pathway. The invention also provides a method of using such status information to select patients responsive to pathway-specific drugs, and more particularly, to methods for measuring ErbB receptors and receptor complexes and using such information to select patients responsive to ErbB pathway-specific drugs. Preferably, methods of the invention are implemented by using sets of binding compounds having releasable molecular tags that are specific for multiple components of one or more complexes formed in RTK activation. After binding, molecular tags are released and separated from the assay mixture for analysis.
Owner:MONOGRAM BIOSCIENCES
Receptor tyrosine kinase signaling pathway analysis for diagnosis and therapy
ActiveUS20050131006A1BiocideMicrobiological testing/measurementPathway analysisProtein-protein complex
The invention provides a method for determining the activation status of receptor tyrosine kinase (RTK) pathways in either cell samples or patient samples by measuring receptor dimerization and relative amounts of protein-protein complexes or activated effector proteins that are characteristic of an RTK pathway. The invention also provides a method of using such status information to select patients responsive to pathway-specific drugs, and more particularly, to methods for measuring ErbB receptors and receptor complexes and using such information to select patients responsive to ErbB pathway-specific drugs. Preferably, methods of the invention are implemented by using sets of binding compounds having releasable molecular tags that are specific for multiple components of one or more complexes formed in RTK activation. After binding, molecular tags are released and separated from the assay mixture for analysis.
Owner:MONOGRAM BIOSCIENCES
Metabolite identification and disorder pathway analysis method
ActiveCN107729721AExtended identification coverageData visualisationMaterial analysis by electric/magnetic meansMetabolitePathway analysis
The invention discloses a metabolite identification and disorder metabolic pathway analysis method. According to the metabolite identification method, the similar structure characteristics and the reaction relationship between a first metabolite and a second metabolite in a metabolic reaction network are used, wherein the reaction relationship exists between the first metabolite and the second metabolite; the second metabolite having the reaction relationship with the first metabolite is identified with a secondary spectrogram of the identified first metabolite, a new second metabolite havingthe reaction relationship with the second metabolite is further identified with a secondary spectrogram of the identified second metabolite. The metabolite identification and disorder metabolic pathway analysis method is circularly carried out till new second metabolites cannot be identified, and therefore the identification covering range of the metabolite is widened; in the disorder metabolic pathway analysis method, through the characteristic that metabolites with the reaction relationship are collected in the metabolic reaction network, the influence of false annotation on pathway analysisis avoided, and disorder network and disorder metabolic pathway information is directly obtained.
Owner:SHANGHAI INST OF ORGANIC CHEM CHINESE ACAD OF SCI
Method for screening and annotating of longissimus dorsi differential expression genes of pigs of different varieties
InactiveCN104636638ADeepen understandingSpecial data processing applicationsLongissimus dorsiLongissimus
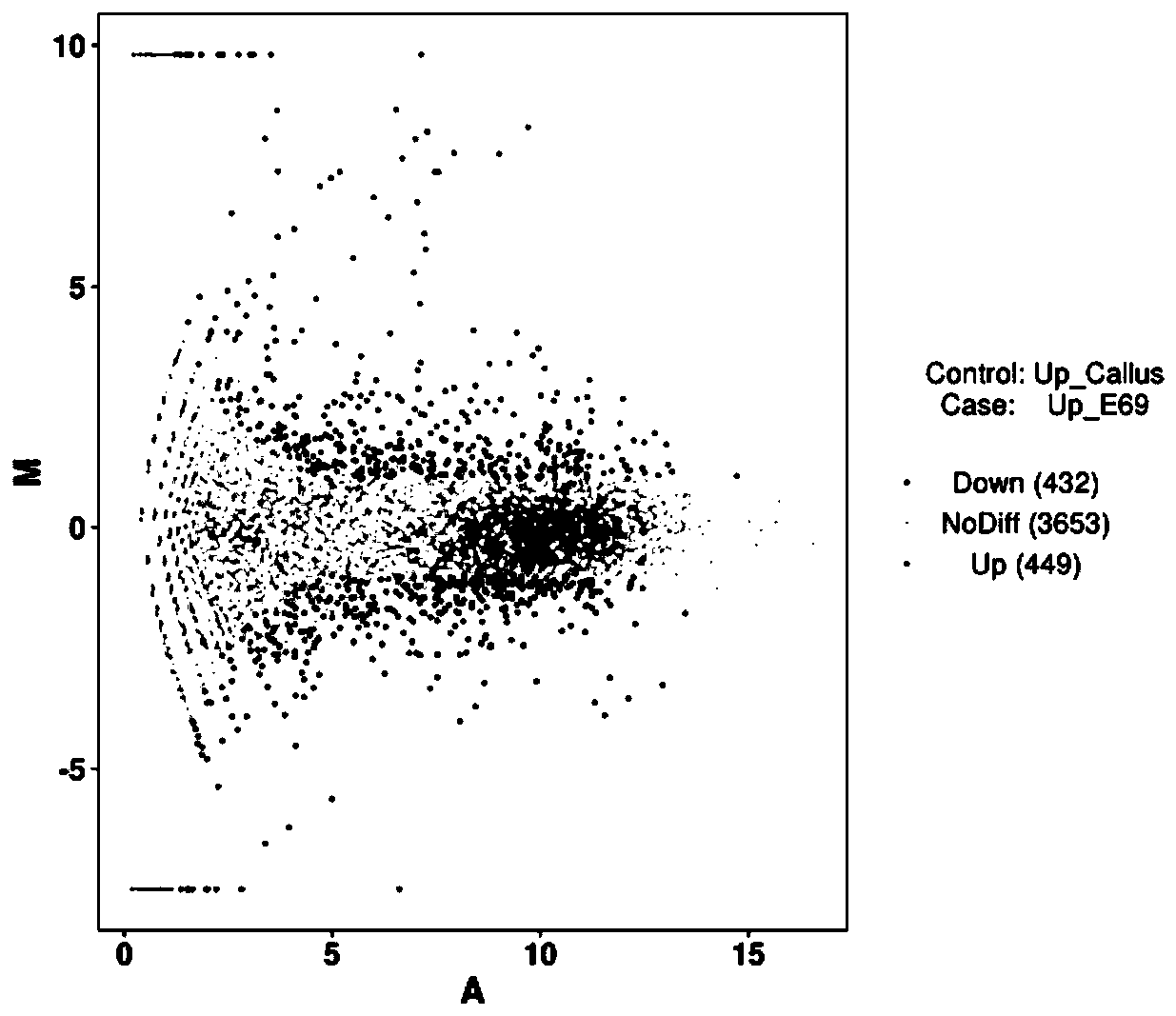
The invention discloses a method for screening and annotating of longissimus dorsi differential expression genes of pigs of different varieties. The method comprises the following steps that 1, sampling and sequencing are conducted; 2, original data are processed; 3, differential genes are screened; 4, function significance enrichment analysis is conducted; 5, Pathway significance enrichment analysis is conducted. According to the method, a high throughout transcriptome sequencing technology is adopted, the gene expression profiles of longissimus dorsi tissue of the pigs of different varieties are established, the differential expression genes are obtained, the differential expression genes are selected through GO analysis and pathway analysis, the key gene of controlling the meat quality trait is determined, control over muscle growth and development and fat metabolism can be better understood, and the molecular biology evidence is provided for the following research of the molecular mechanism forming the difference of the meat quality and the growth trait of domestic and oversea varieties.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Biological fermented feed and preparation method thereof
ActiveCN106376725AAdvanced technologyReduce human transportationFungiBacteriaFiberBacillus licheniformis
The invention relates to biological feed, in particular to a biological fermented feed and a preparation method thereof. The method consists of: subjecting Aspergillus niger, Bacillus licheniformis, Bacillus coagulans, Bacillus subtilis, Lactobacillus plantarum, and Candida utilis respectively to two-stage liquid fermentation culture, and then conducting three-stage solid fermentation culture to obtain fermented culture, i.e. biological feed. The fermented culture provided by the invention utilizes neural network, pattern recognition, genetic algorithm and other optimization methods to optimize the fermentation conditions, a biosynthesis network is constructed according to the principle of metabolic engineering, and pathway analysis and metabolic flux control are carried out on the synthetic network on the basis, and enzymolysis, anaerobism, aerobiosis, bacterium cultivation and other processes are employed, thus increasing the protein content, reducing the content of crude fiber, improving the palatability and fragrance, increasing probiotics, and removing toxins from feed raw materials.
Owner:北京康缘益生生物科技有限公司
Method for analyzing proteins contributing to autoimmune diseases, and method for testing for said diseases
ActiveUS20130273579A1High sensitivityImprove efficiencyDisease diagnosisBiological testingImmunologic disordersDisease
Provided are a detection method for a myriad of proteins involved in an autoimmune disease with high sensitivity and high efficiency, and an analysis method for data resulting from the detection method. In order to construct the detection method and analysis method, there is provided means for comprehensively analyzing the proteins involved in an autoimmune disease by bringing a mammal-derived protein expressed in a cell-free protein synthesis system into contact with a sample derived from a patient with an autoimmune disease to detect autoantibody production, and subjecting the detected data to statistical analysis processing, and further, gene ontology analysis and / or pathway analysis.
Owner:PUBLIC UNIV CORP YOKOHAMA CITY UNIV
Identification of esophageal cancer related characteristic pathways and construction method of early diagnosis model
ActiveCN109841280ALimit random fluctuationsFine molecular mechanismHealth-index calculationMedical automated diagnosisPathway analysisWilms' tumor
The invention belongs to the technical field of tumor diagnosis, and particularly relates to identification of esophageal cancer related characteristic pathways of and a construction method of an early diagnosis model. The identification and constructing method includes the steps: expression spectrum preprocessing, differential expression gene extraction, sample cluster analysis, gene cluster analysis, specific gene set functional pathway analysis, comparison of pathway aberration scores, comparative analysis of functional differences, construction of esophageal cancer specific co-expression networks, characteristic selection of genes, prediction of deep learning models, and the like. The method of the invention divides genes into different groups according to expression similarity and functional consistency of the genes, and analyzes the genes in the form of gene collection, thus being able to avoid the disadvantages of high false positive rate, big random error and unstable result inthe traditional method, and also being able to more specifically identify the function significantly associated with esophageal cancer.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
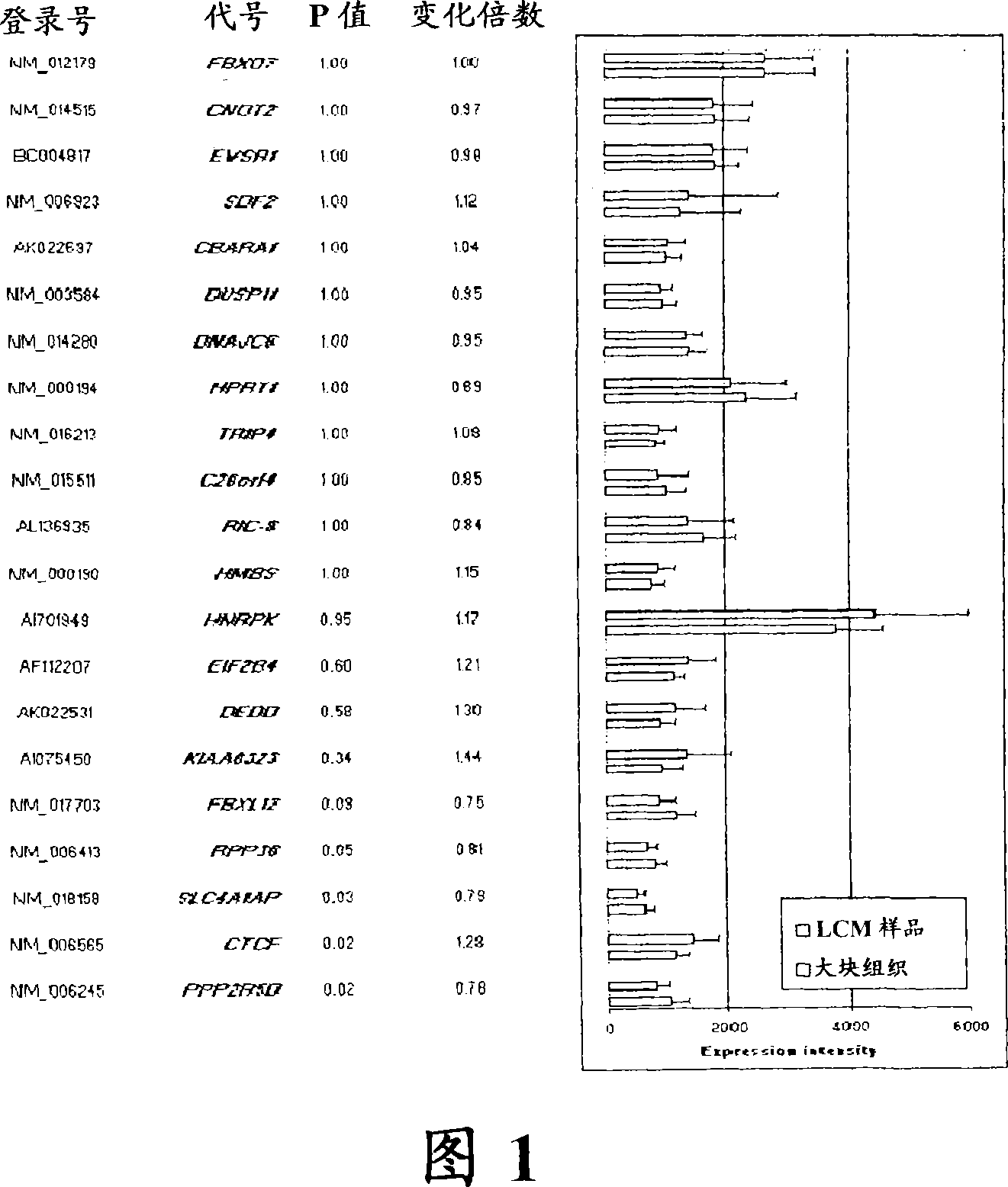
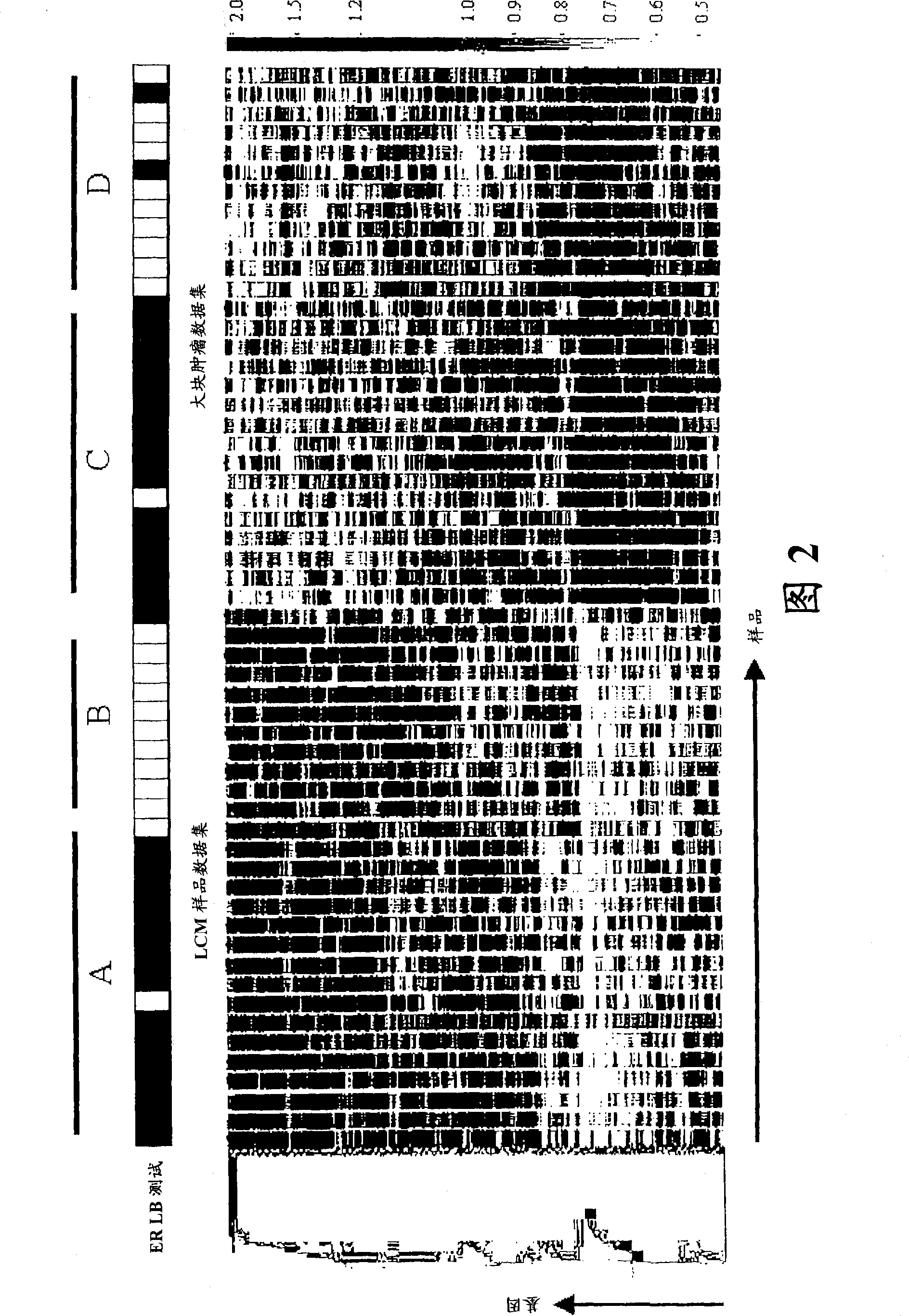
Laser microdissection and microarray analysis of breast tumors reveal estrogen receptor related genes and pathways
InactiveCN101965190APeptide/protein ingredientsMicrobiological testing/measurementPathway analysisLaser micro dissection
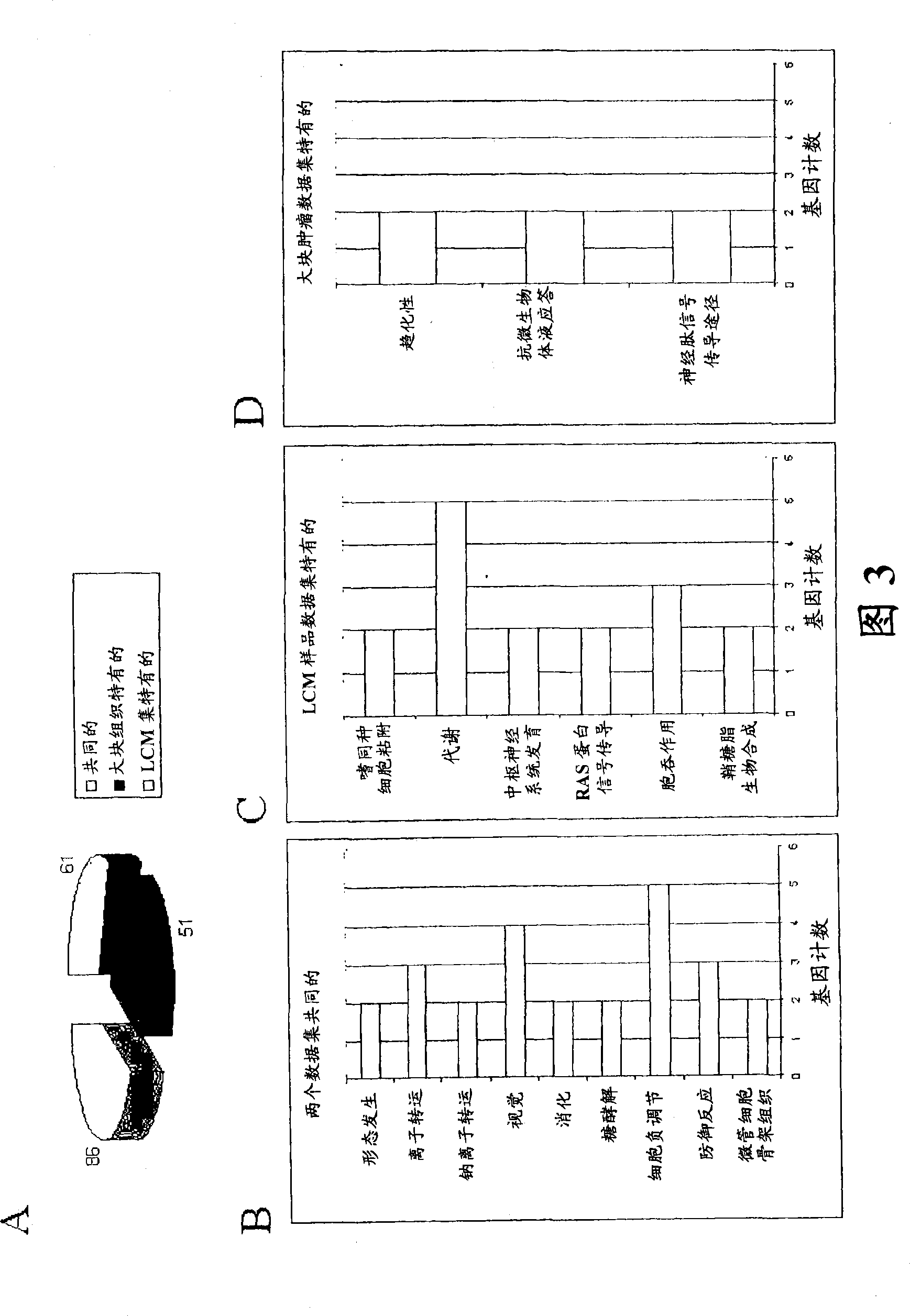
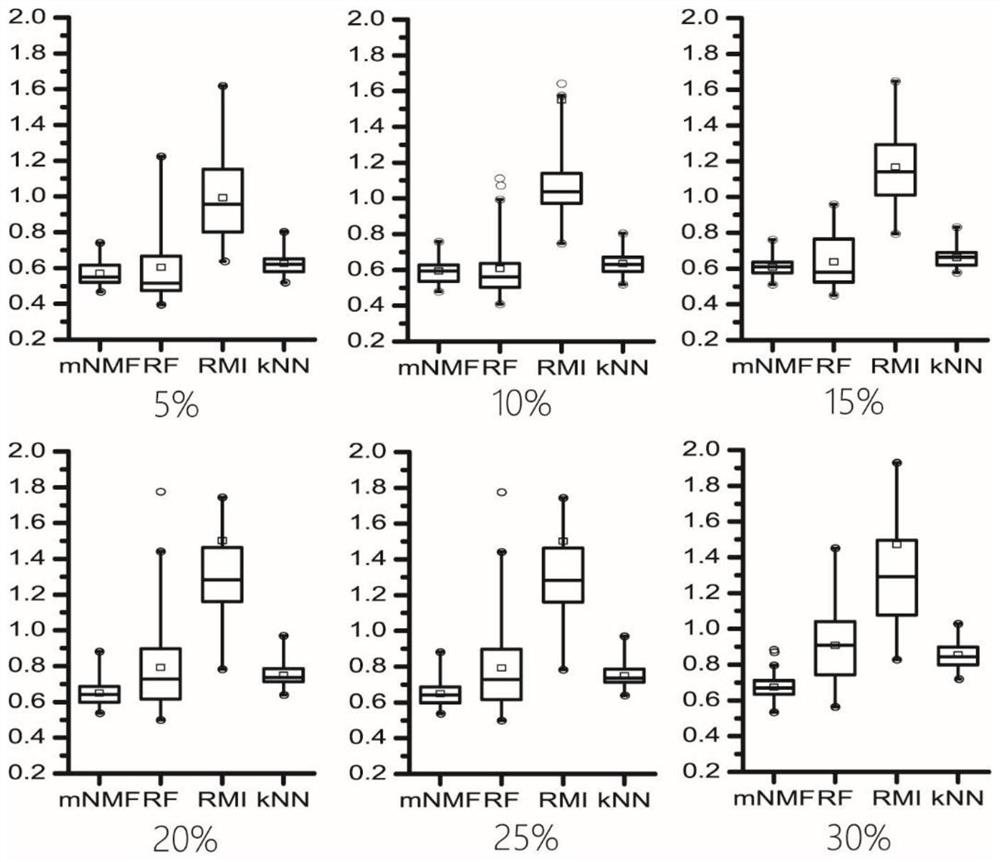
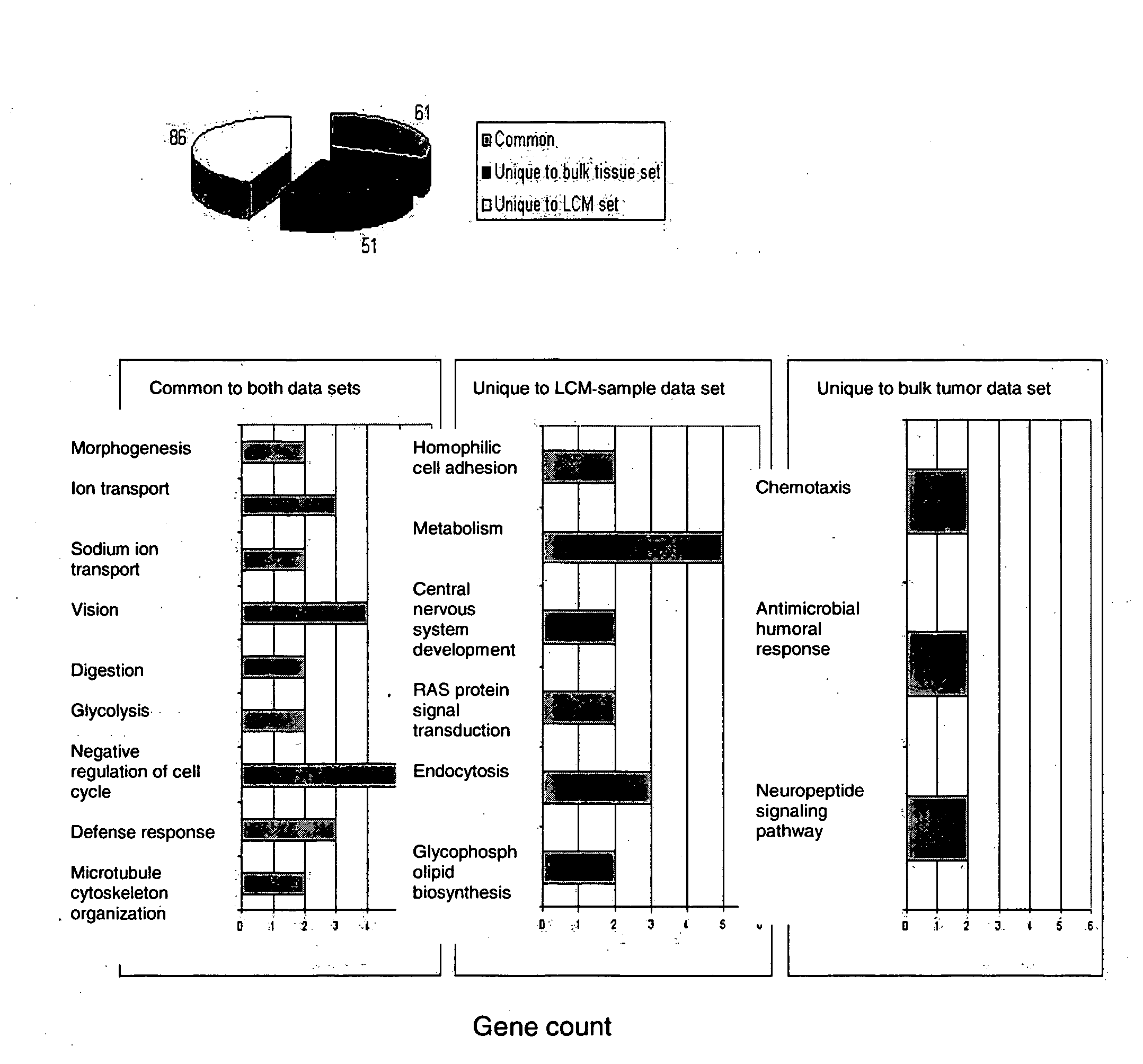
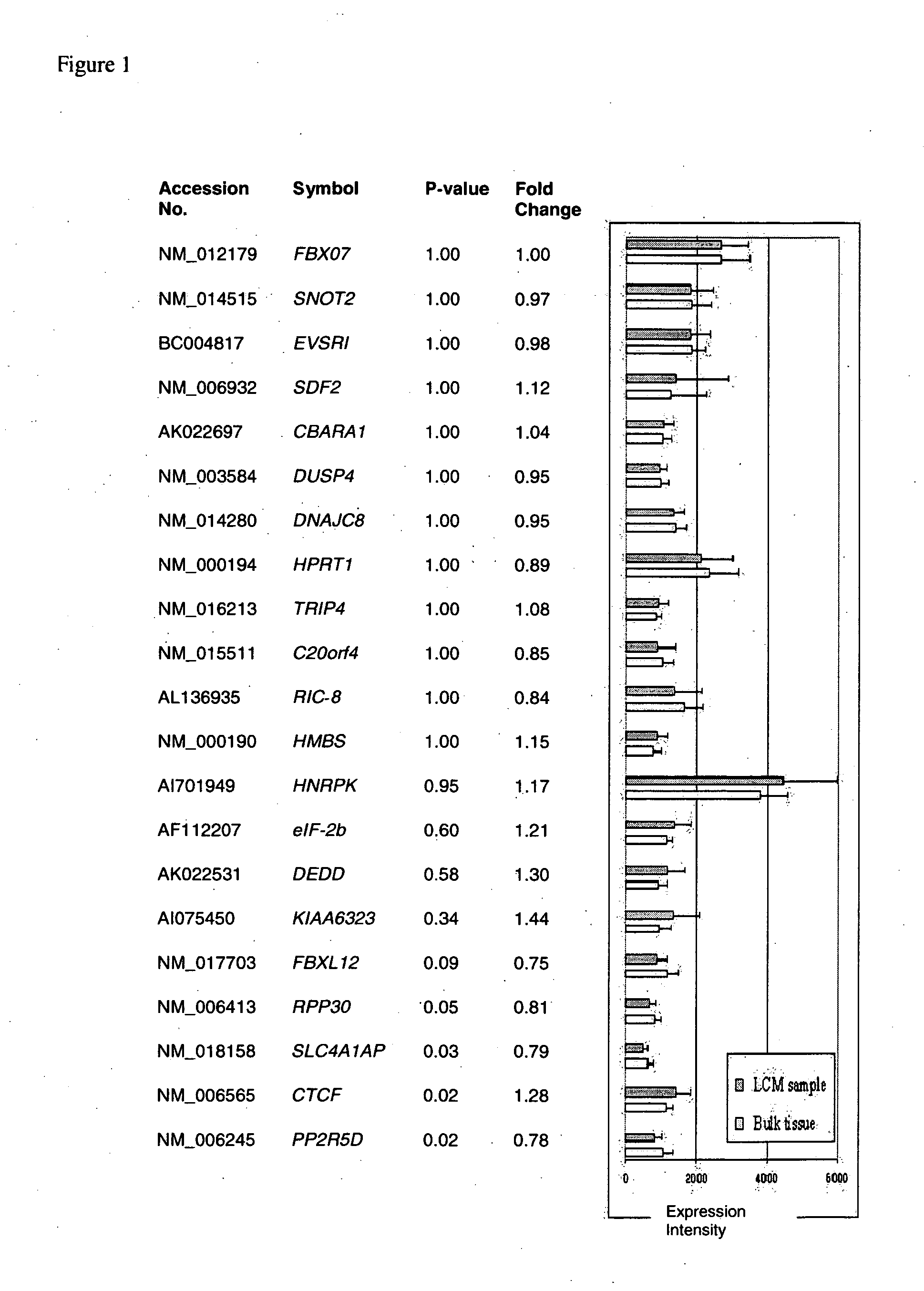
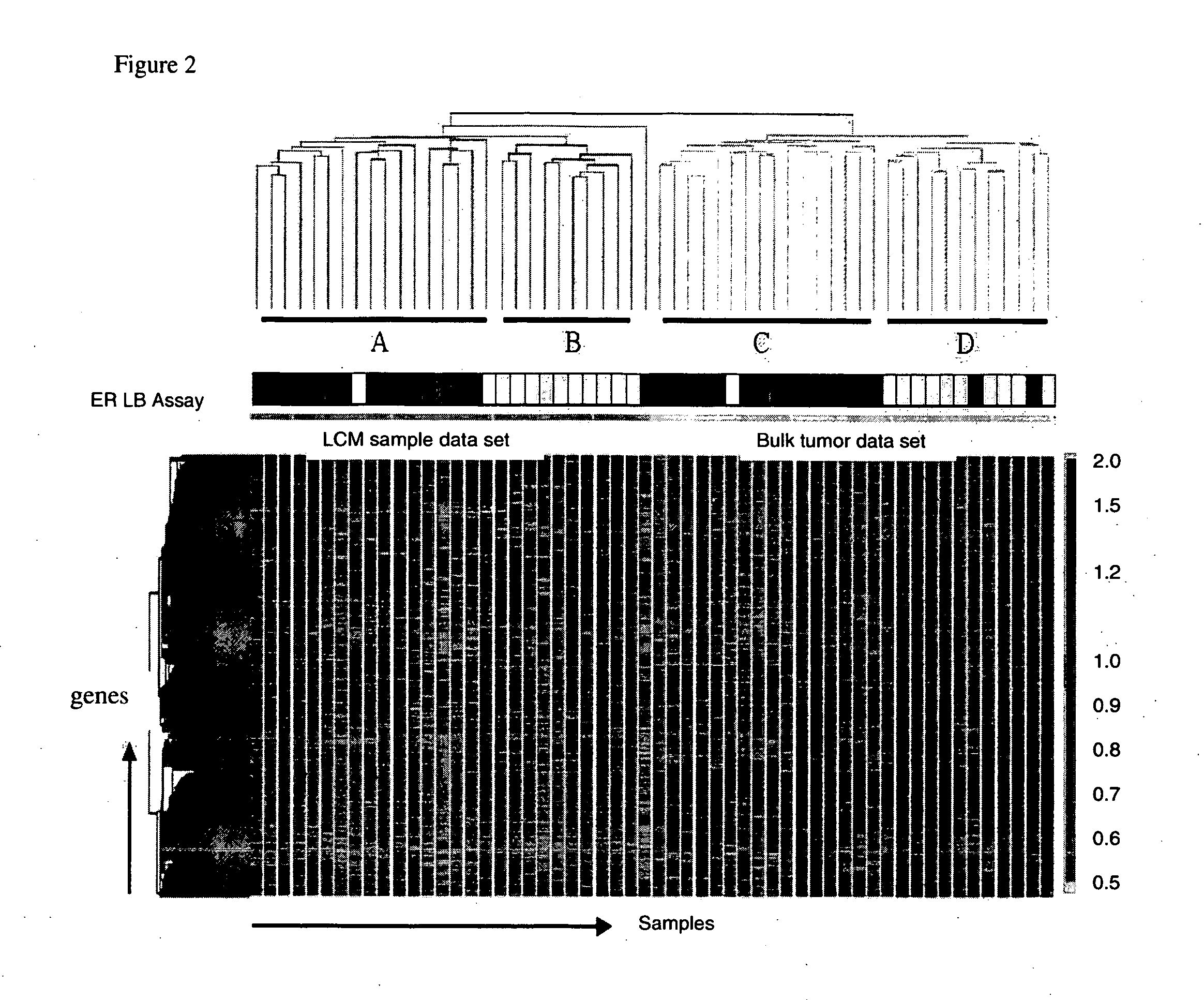
About 70% to 80% of breast cancers express estrogen receptor-a (ERa), and estrogens play important roles in the development and growth of hormone-dependent tumors. Together with lymph node metastasis, tumor size and histological grade, ER status is considered one of the prognostic factors in breast cancer, and an indicator for hormonal treatment. 147 genes and 112 genes with significant P- value and having significant differential expression between ER+ and ER- tumors were identified from the LCM data set and bulk tissue data set, respectively. 61 genes were found to be common in both data sets, while 85 genes were unique to the LCM data set and 51 genes were present only in the bulk tumor data set. Pathway analysis with the 85 genes using Gene Ontology suggested that genes involved in endocytosis, ceramide generation, Ras / ERK / Ark cascade, and JAT- STAT pathway may play roles related to ER. The gene profiling with LCM-captured tumpr cells provides a unique approach to characterize and study epithelial tumor cells and to gain an insight into signaling pathways associated with ER.
Owner:VERIDEX LCC
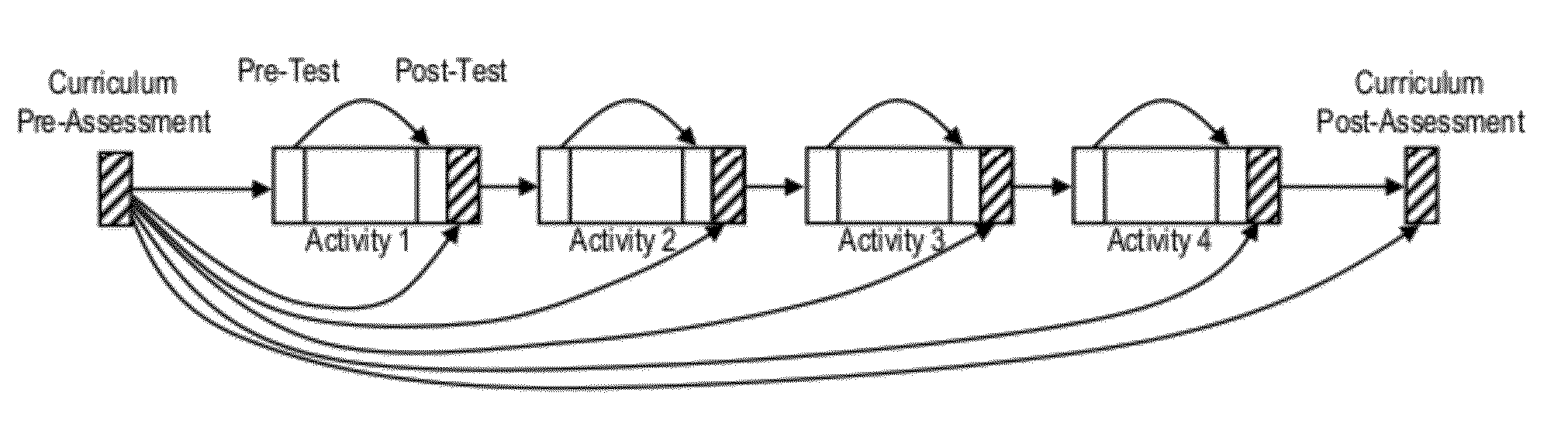
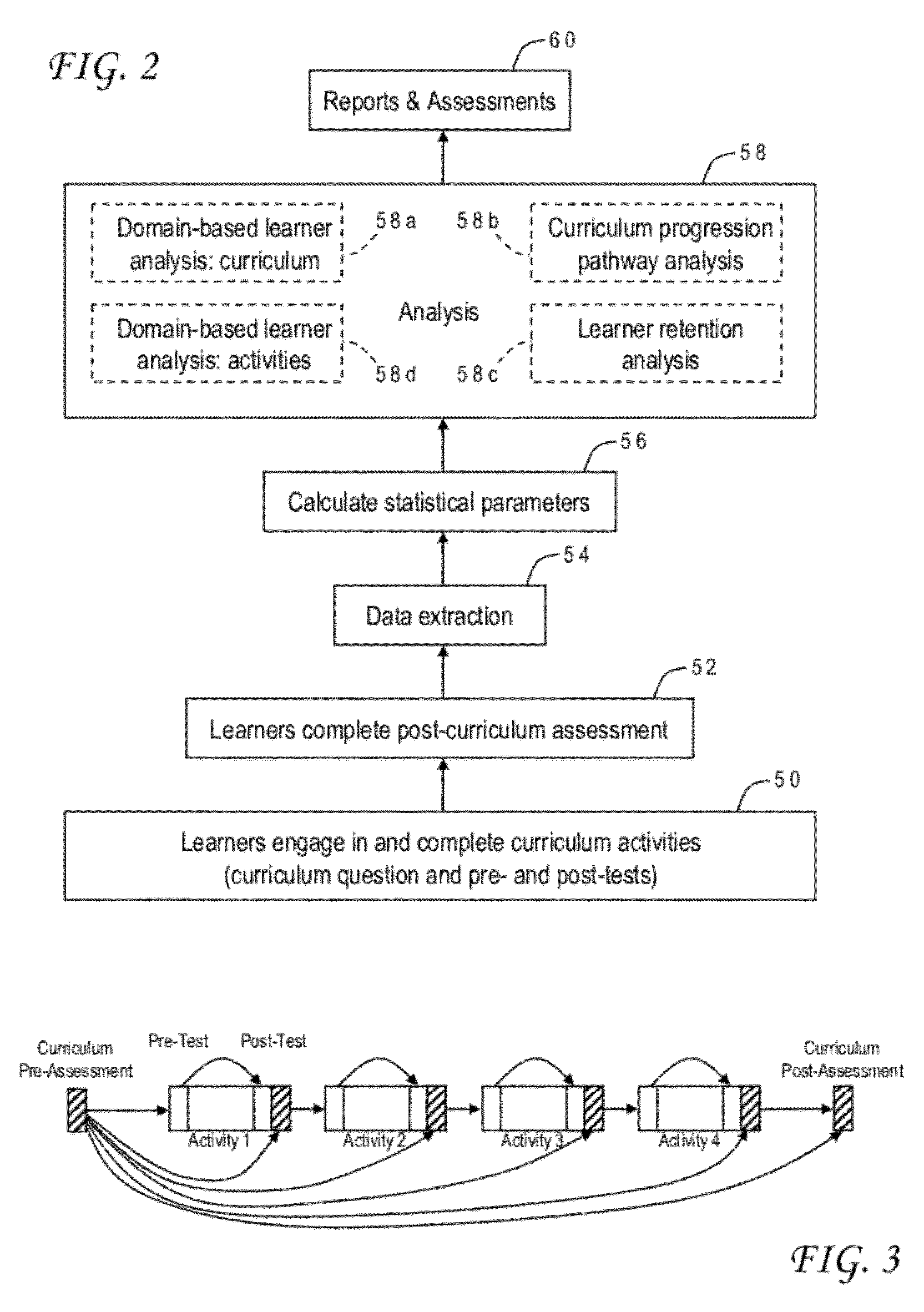
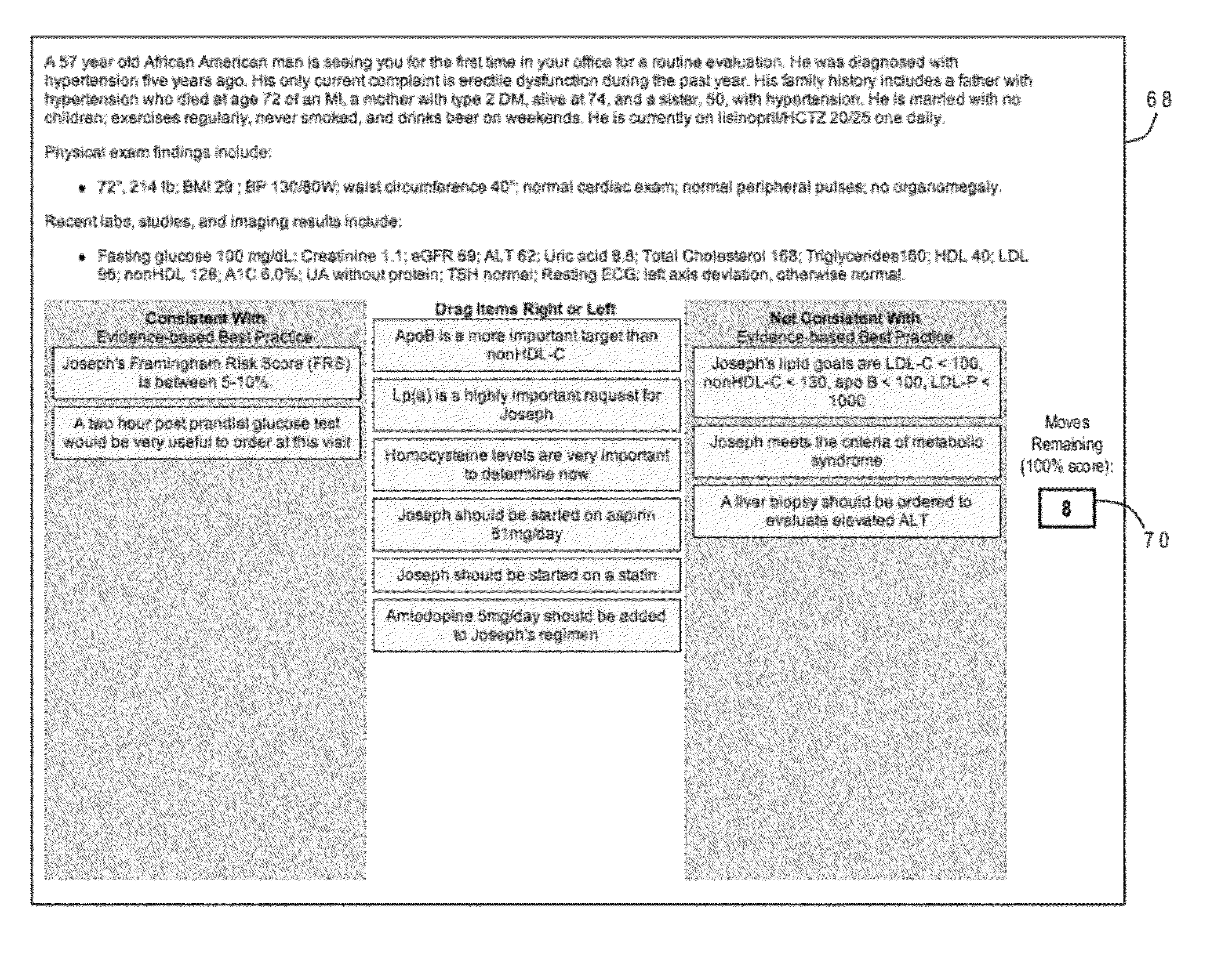
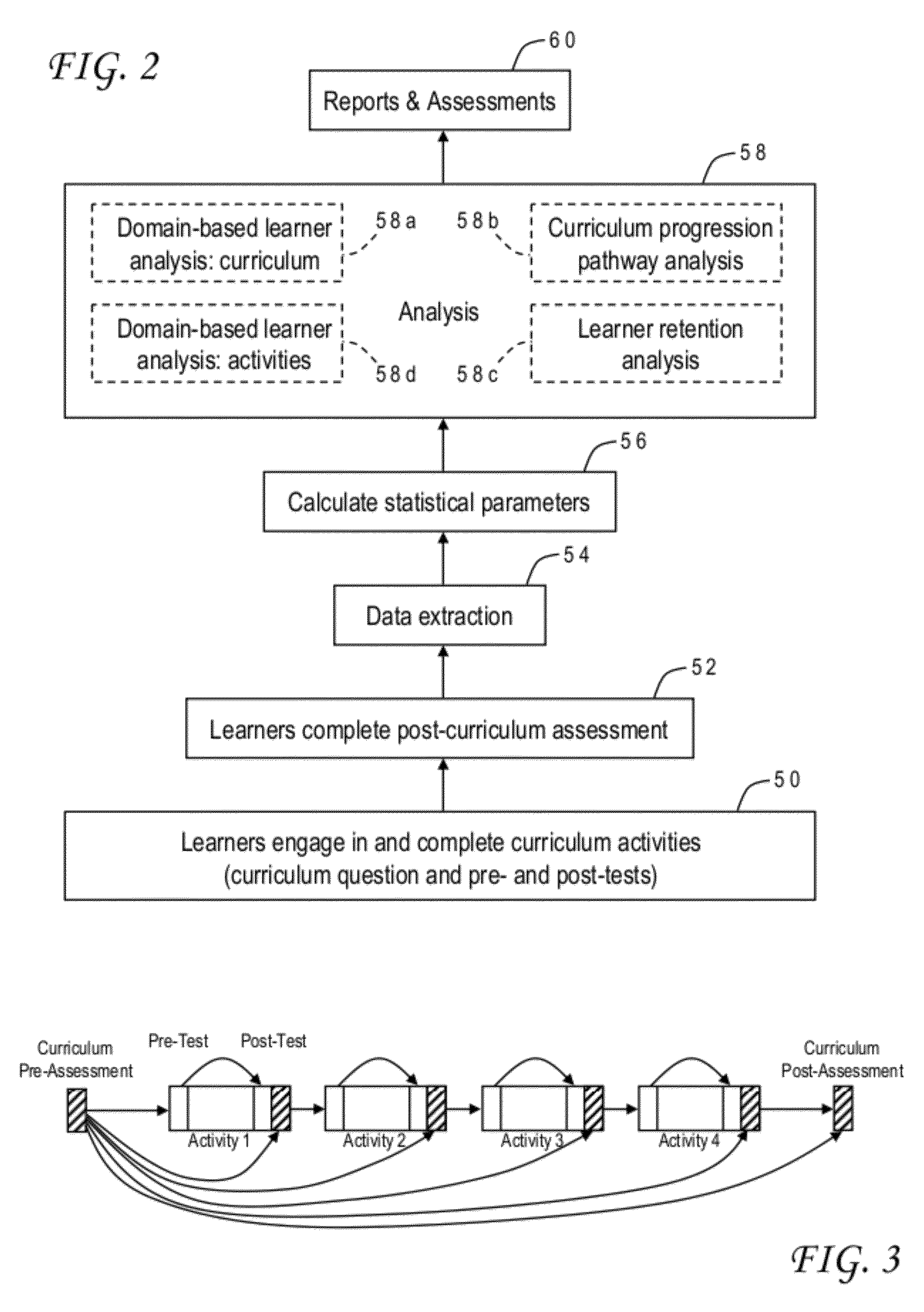
Educational program assessment using curriculum progression pathway analysis
ActiveUS20120276514A1Simple methodElectrical appliancesMechanical appliancesPathway analysisPerformance index
A curriculum progression pathway analysis assesses an educational program having multiple activities defining multiple learner tracks by organizing the learners into groups based on the learner tracks and, for each learner group, calculating a baseline average score, a post-activity average score, and statistical differences between the baseline average score and the post-activity average scores. Any significant statistical differences are identified, and a report may be generated describing statistical conclusions and inferences. The invention can further provide a domain-based learner analysis for curriculum, a domain-based learner analysis for activities, and a learner retention analysis. In an application for continuing medical education the performance index test includes a clinical vignette, a set of statements which are moved to either a best practices column or a not best practices column, and an indicator for an allowable number of moves to achieve a perfect score.
Owner:REALCME
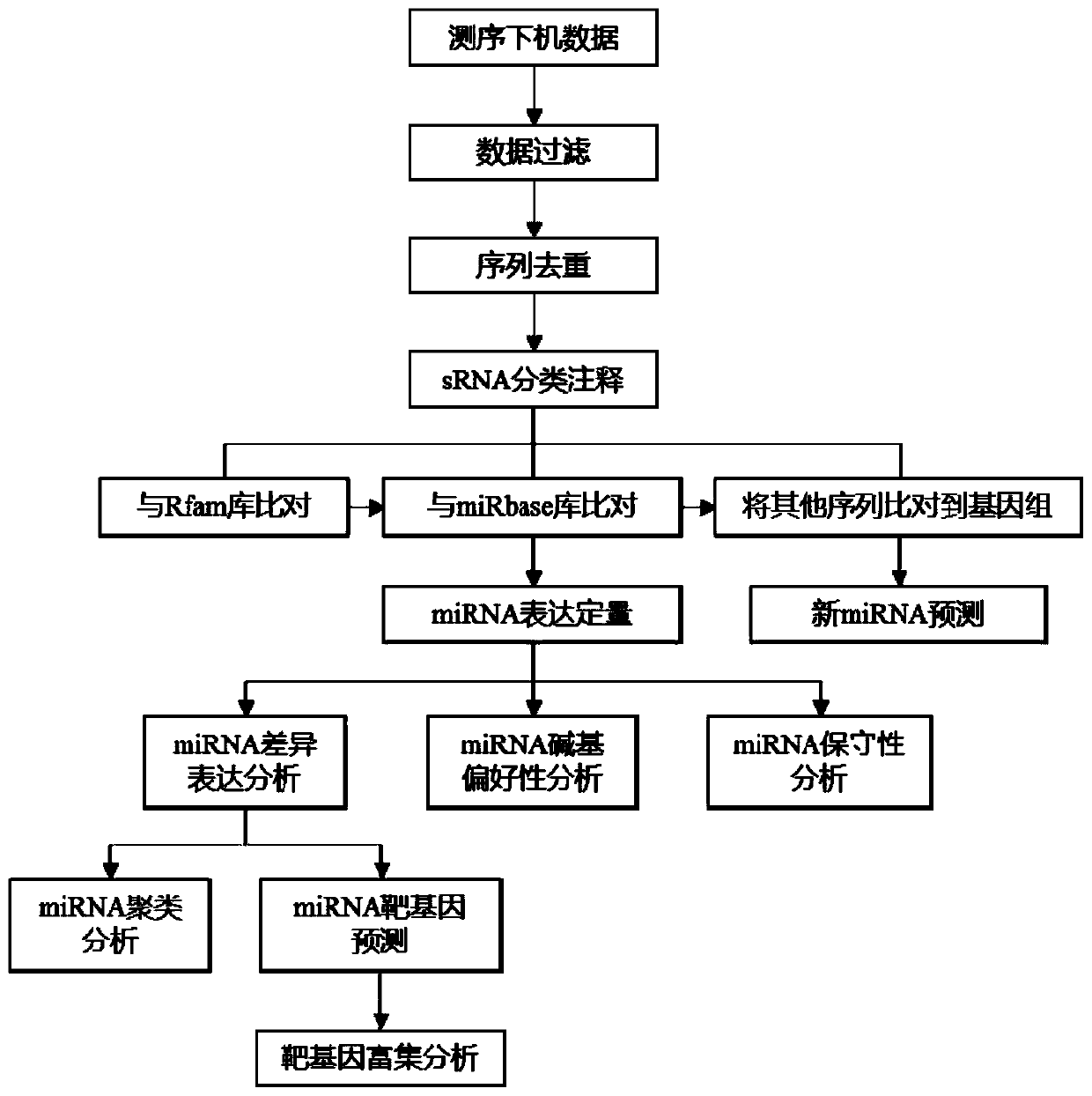
Non-parametric miRNA data analysis method based on miRBase database
The invention discloses a non-parametric miRNA data analysis method based on a miRBase database. The non-parametric miRNA data analysis method includes the following steps: a file preparation step; alogout data filtering step; a sRNA classified annotation step; a miRNA differential analysis step; a miRNA function and pathway analysis step; a miRNA sequence feature analysis step; and a result arrangement step. The non-parametric miRNA data analysis method based on a miRBase database has the advantages of performing analysis of miRNA sequencing data for reference-free miRNA sequences without the species; being comprehensive in the results including the miRNA analysis content involved and other measured small RNA information annotations; automatically arranging all the analysis results, andafter completing the analysis of each part, automatically counting the results, being visualized, and performing sorting and arrangement to enable the results to be arranged at a glance and used directly for report generation; and being visible for all the steps and being easy to query the errors.
Owner:南京派森诺基因科技有限公司
Compositions for cancer treatment and methods and uses for cancer treatment and prognosis
InactiveUS20200078401A1Strong cytotoxicityIncreased proliferationMicrobiological testing/measurementMammal material medical ingredientsPathway analysisTranscriptional analysis
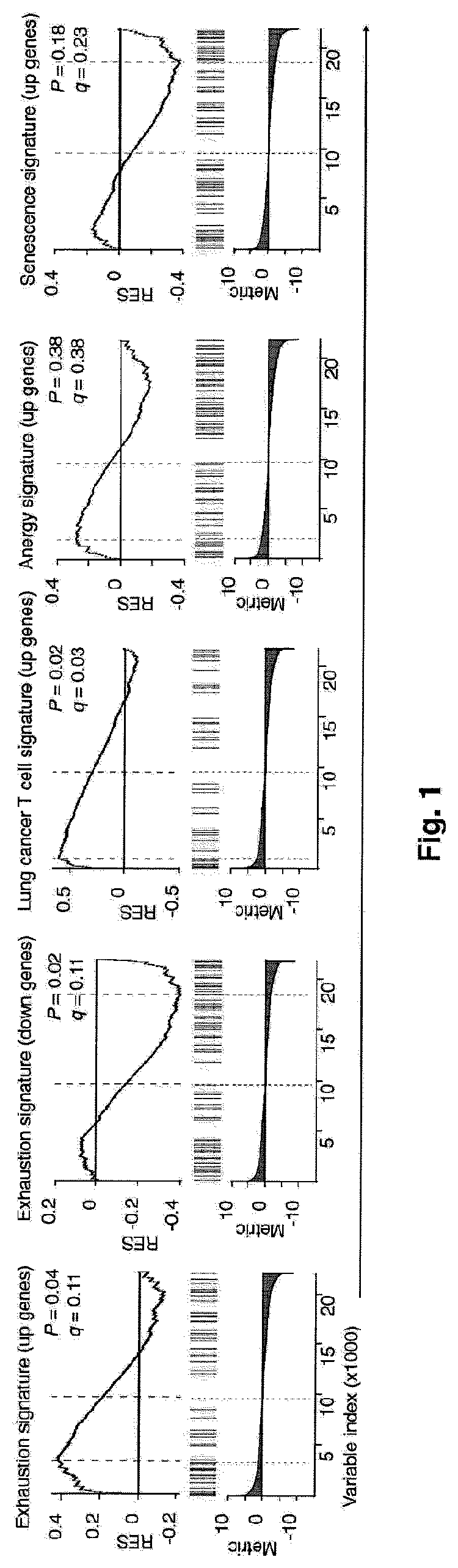
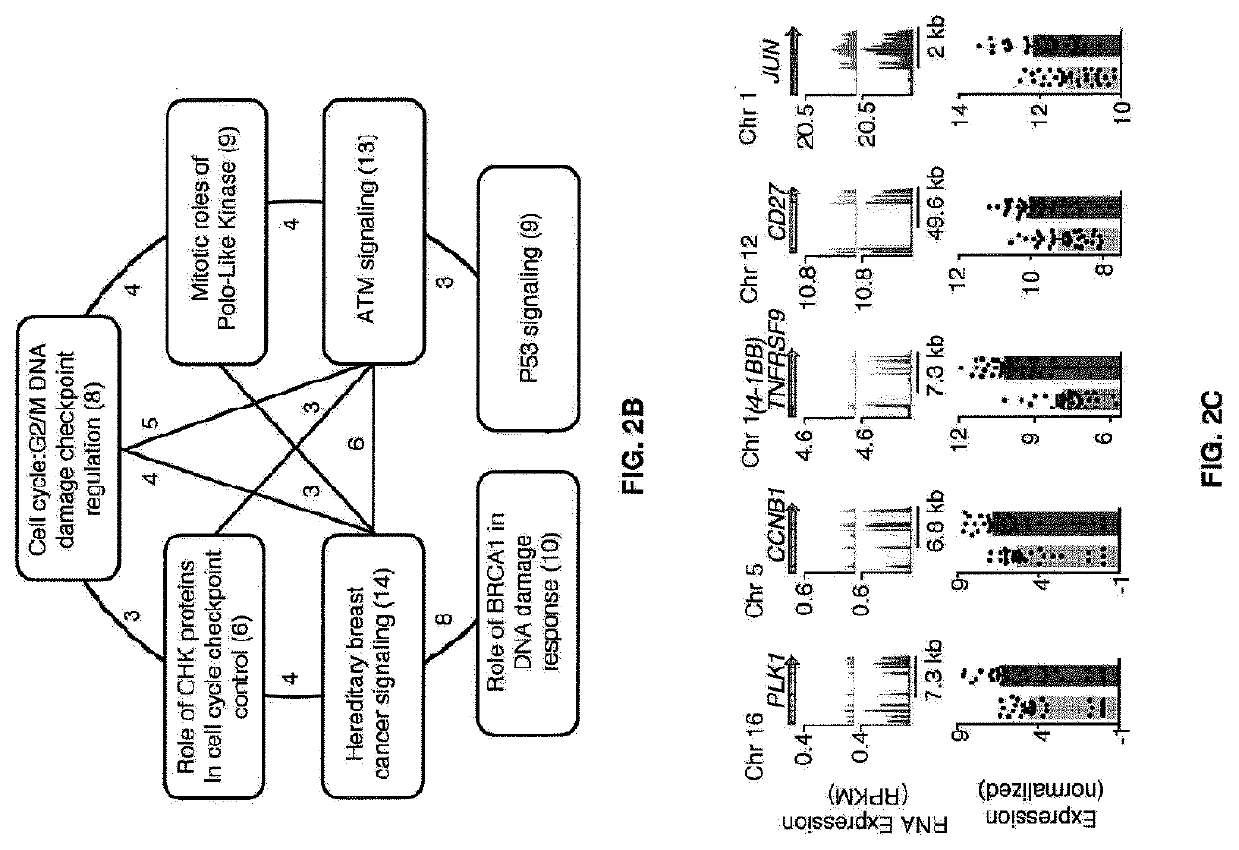
Global transcriptional profiling of CTLs in tumors and adjacent non-tumor tissue from treatment-naive patients with early stage lung cancer revealed molecular features associated with robustness of anti-tumor immune responses. Major differences in the transcriptional program of tumor-infiltrating CTLs were observed that are shared across tumor subtypes. Pathway analysis revealed enrichment of genes in cell cycle, T cell receptor (TCR) activation and co-stimulation pathways, indicating tumor-driven expansion of presumed tumor antigen-specific CTLs. Marked heterogeneity in the expression of molecules associated with TCR activation and immune checkpoints such as 4-1BB, PD1, TIM3, was also observed and their expression was positively correlated with the density of tumor-infiltrating CTLs. Transcripts linked to tissue-resident memory cells (TRM), such as CD 103, were enriched in tumors containing a high density of CTLs, and CTLs from CD 103high tumors displayed features of enhanced cytotoxicity, implying better anti-tumor activity. In an independent cohort of 689 lung cancer patients, patients with CD103high (TRM rich) tumors survived significantly longer. In summary, the molecular fingerprint of tumor-infiltrating CTLs at the site of primary tumor was defined and a number of novel targets identified that appear to be important in modulating the magnitude and specificity of anti-tumor immune responses in lung cancer.
Owner:LA JOLLA INST FOR ALLERGY & IMMUNOLOGY +1
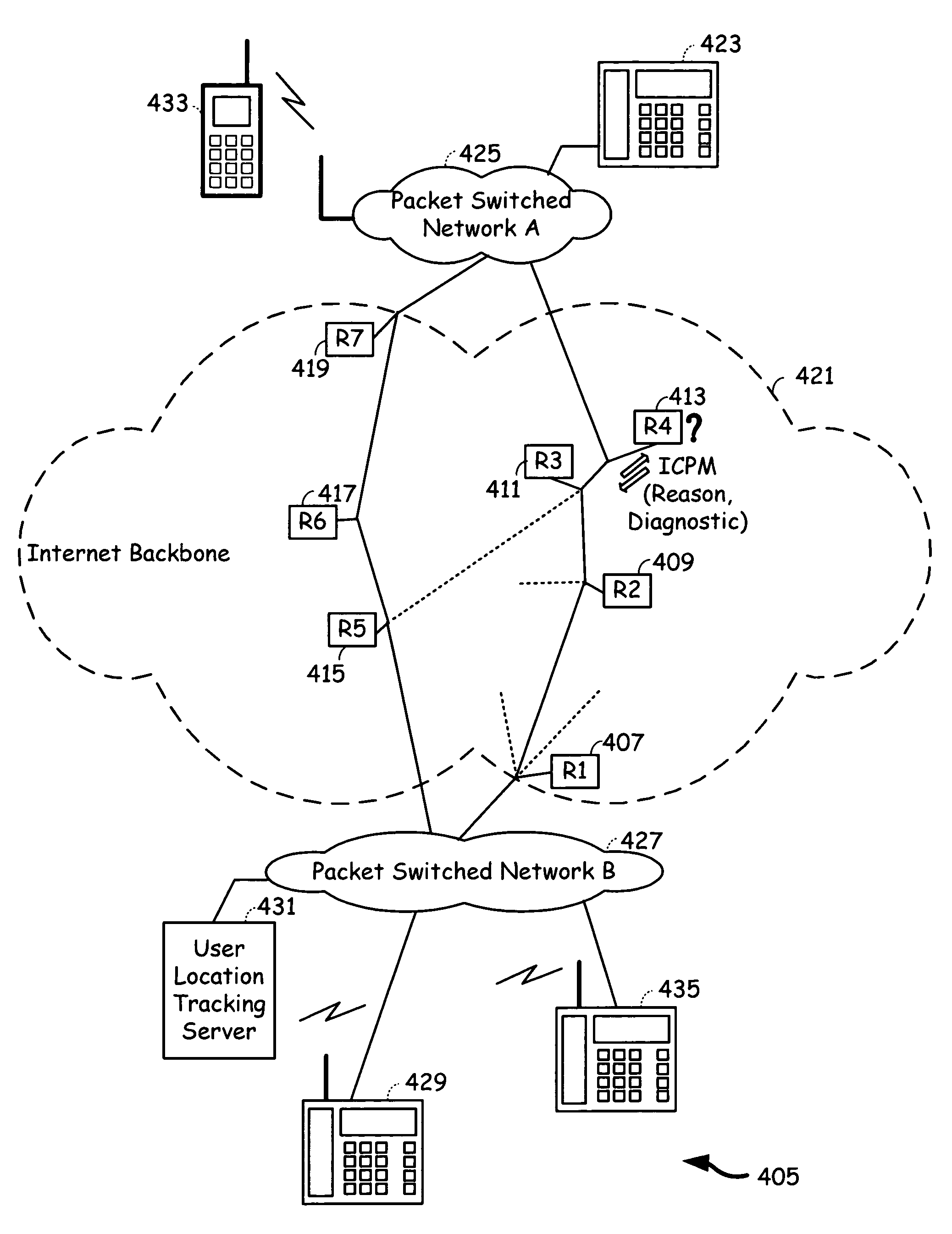
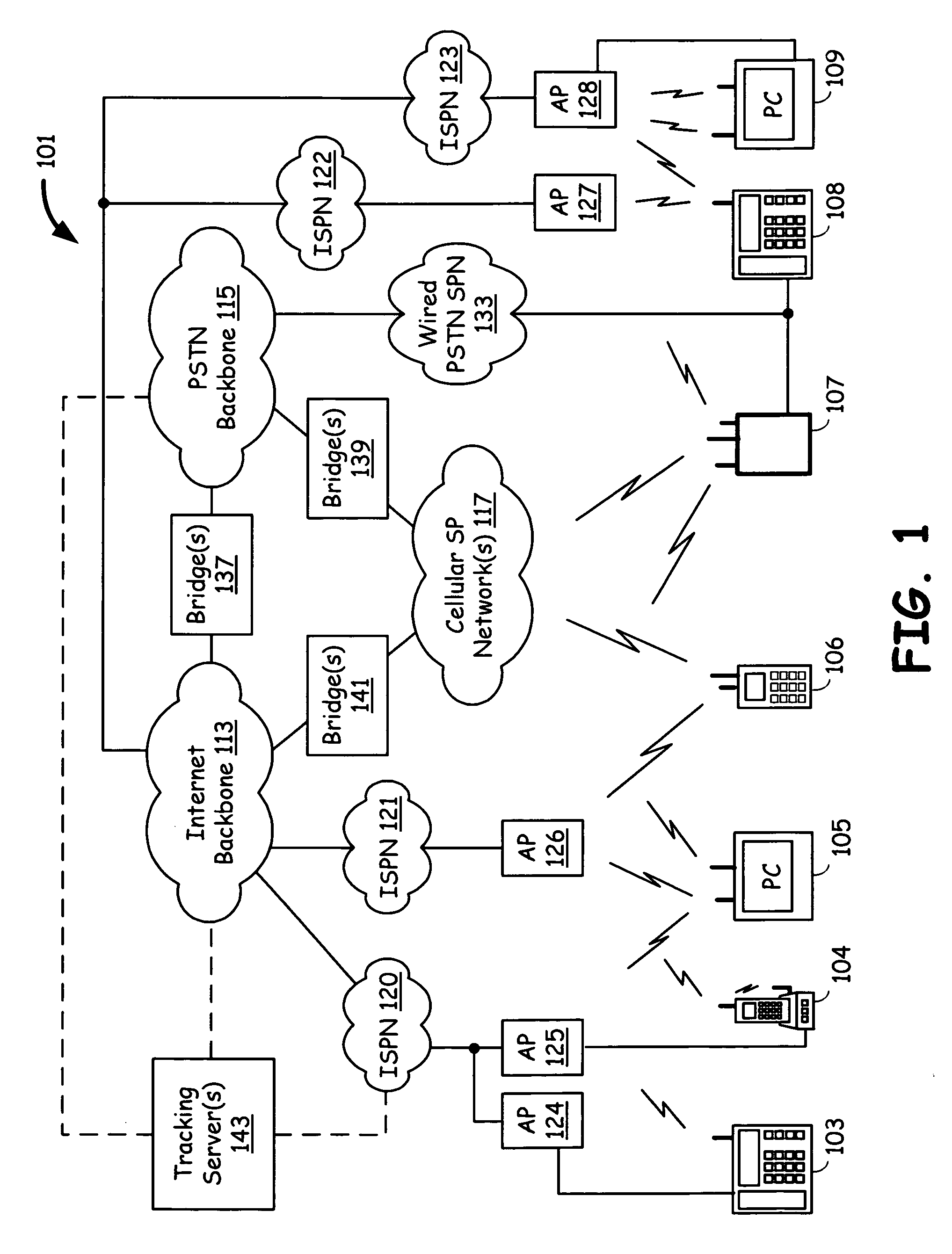
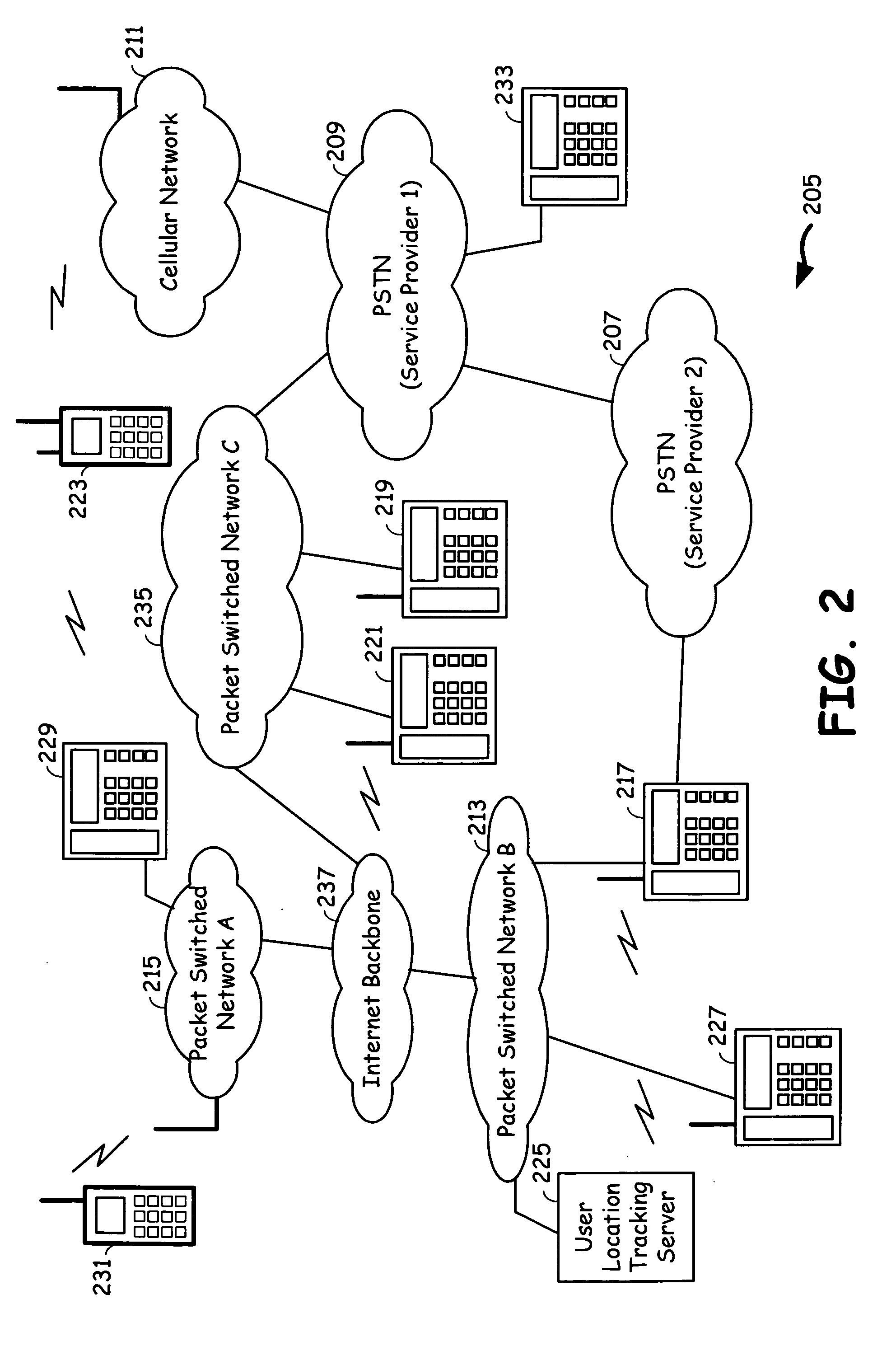
Voice communication device with PSTN and internet pathway analysis, selection and handoff
ActiveUS8274970B2Time-division multiplexData switching by path configurationQuality of serviceCommunication interface
A voice communication device that supports both Internet telephony and public switched telephone network telephony, and maintains the quality of communication. The voice communication device consists of a user interface, a plurality of communication interfaces and a processing circuitry communicatively coupled to the user interface and to the plurality of communication interfaces. The voice communication device maintains quality of service by analyzing the service characteristics of a plurality of communication pathways and determining one communication pathway that compares best in terms of quality of service and cost and transferring to that communication pathway prior to or during the call. Alternatively, a user set service characteristic configurations may also determine the choice of a communication pathway that meets the user requirements of quality of service and cost.
Owner:AVAGO TECH INT SALES PTE LTD
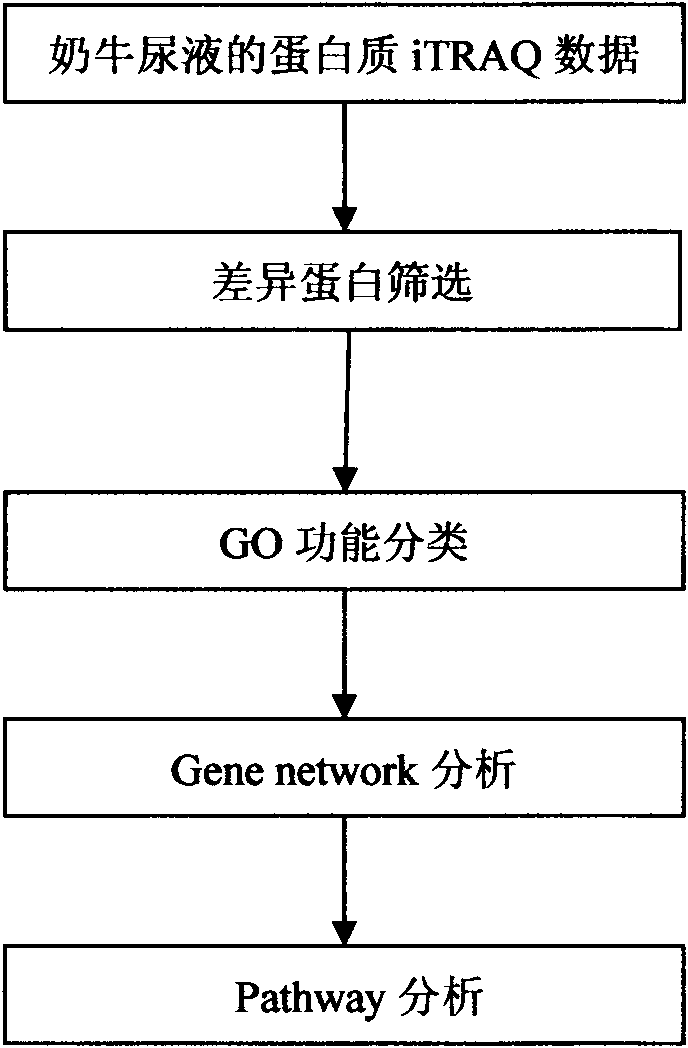
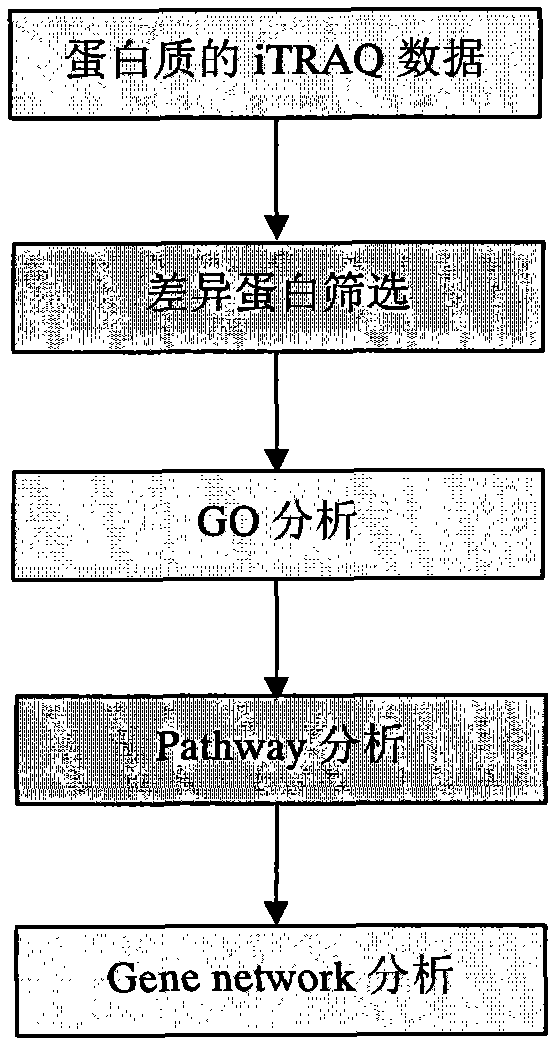
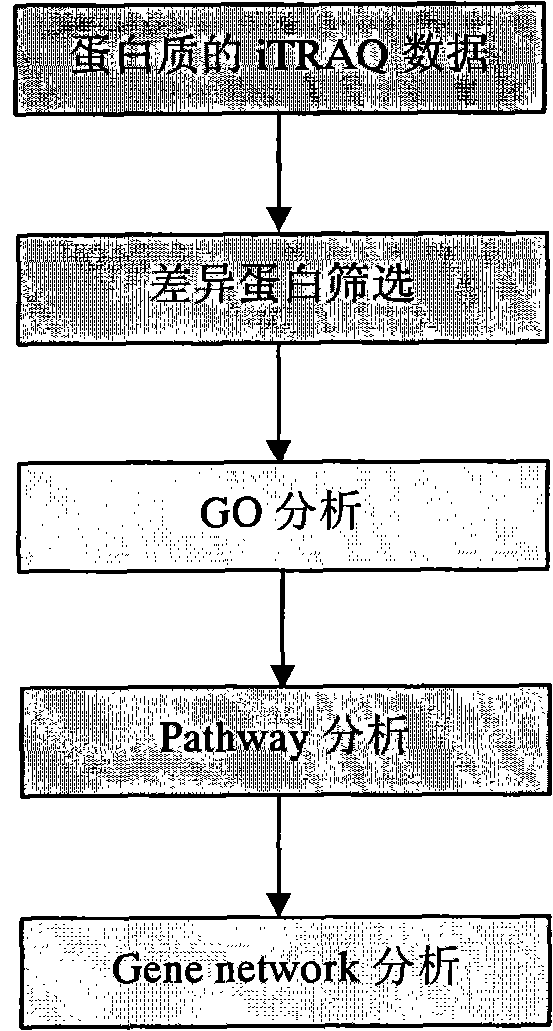
Processing method of cow urine iTRAQ test data
Aiming at the characteristics of iTRAQ protein quantitative analysis, a method for analyzing cow urine iTRAQ data is designed. The main process includes: step one, screening differential proteins and obtaining the differentiated result by grouping and screening the original iTRAQ data; step two, performing GO analysis, including obtaining more comprehensive gene function information displayed as a directed acyclic graph through mapping query to the GO database; step three, performing Pathway analysis, which is similar with the GO analysis, including construction of interaction paths between genes through the comparison and query of the KEGG database; and step four, performing gene network analysis, including obtaining a gene interaction network diagram through the integration of three different types of interactions.
Owner:SHANGHAI CLUSTER BIOTECH
Systems And Methods For Comprehensive Analysis Of Molecular Profiles Across Multiple Tumor And Germline Exomes
Omics patient data are analyzed using sequences or diff objects of tumor and matched normal tissue to identify patient and disease specific mutations, using transcriptomic data to identify expression levels of the mutated genes, and pathway analysis based on the so obtained omic data to identify specific pathway characteristics for the diseased tissue. Most notably, many different tumors have shared pathway characteristics, and identification of a pathway characteristic of a tumor may thus indicate effective treatment options ordinarily not considered when tumor analysis is based on anatomical tumor type only.
Owner:FIVE3 GENOMICS +2
GC-MS (Gas Chromatography-Mass Spectrometer) combined technology-based metabonomics research method for resistance of asarum and red peony root composition to cerebral ischemia-reperfusion injury
The invention provides a GC-MS (Gas Chromatography-Mass Spectrometer) combined technology-based metabonomics research method for resistance of an asarum and red peony root composition to a cerebral ischemia-reperfusion injury. The method is realized through the following steps: (1) detecting and analyzing a serum sample and a urine sample of a mouse by adopting a GC-MS combined technology; (2) analyzing data by using PCA (Primary Component Analysis) and PLS-DA (Partial Least Square)-DA (Discriminant Analysis),and performing metabolite identification in combination with SPSS17.0 by using an NIST (National Institute of Standards and Technology) database to find out a final differential metabolite; (3) performing pathway analysis on the differential metabolite through MetPA online analysis software. According to the method, by analysis of the serum sample and the urine sample of the mouse through the GC-MS combined technology, respective identification of the differential metabolites of the serum sample and the urine sample of the mouse, and metabolic pathway analysis of the differential metabolites, the action mechanism of the asarum and red peony root composition resisting the cerebral ischemia-reperfusion injury can be comprehensively evaluated from the overall level, so that a demonstrative research is provided for explanation of the metabonomics and the action mechanism of a traditional Chinese medicine composition.
Owner:GUIZHOU MEDICAL UNIV
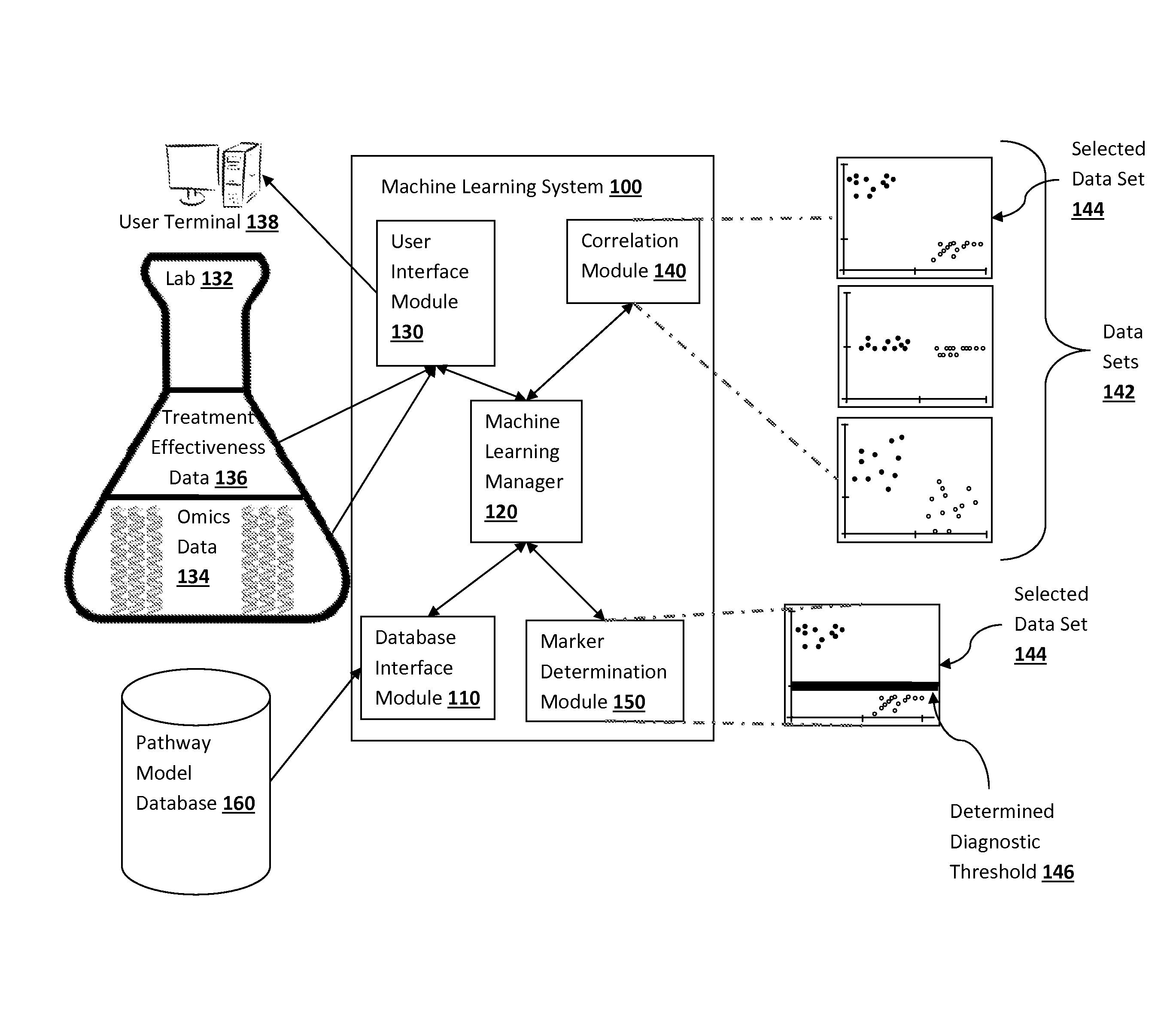
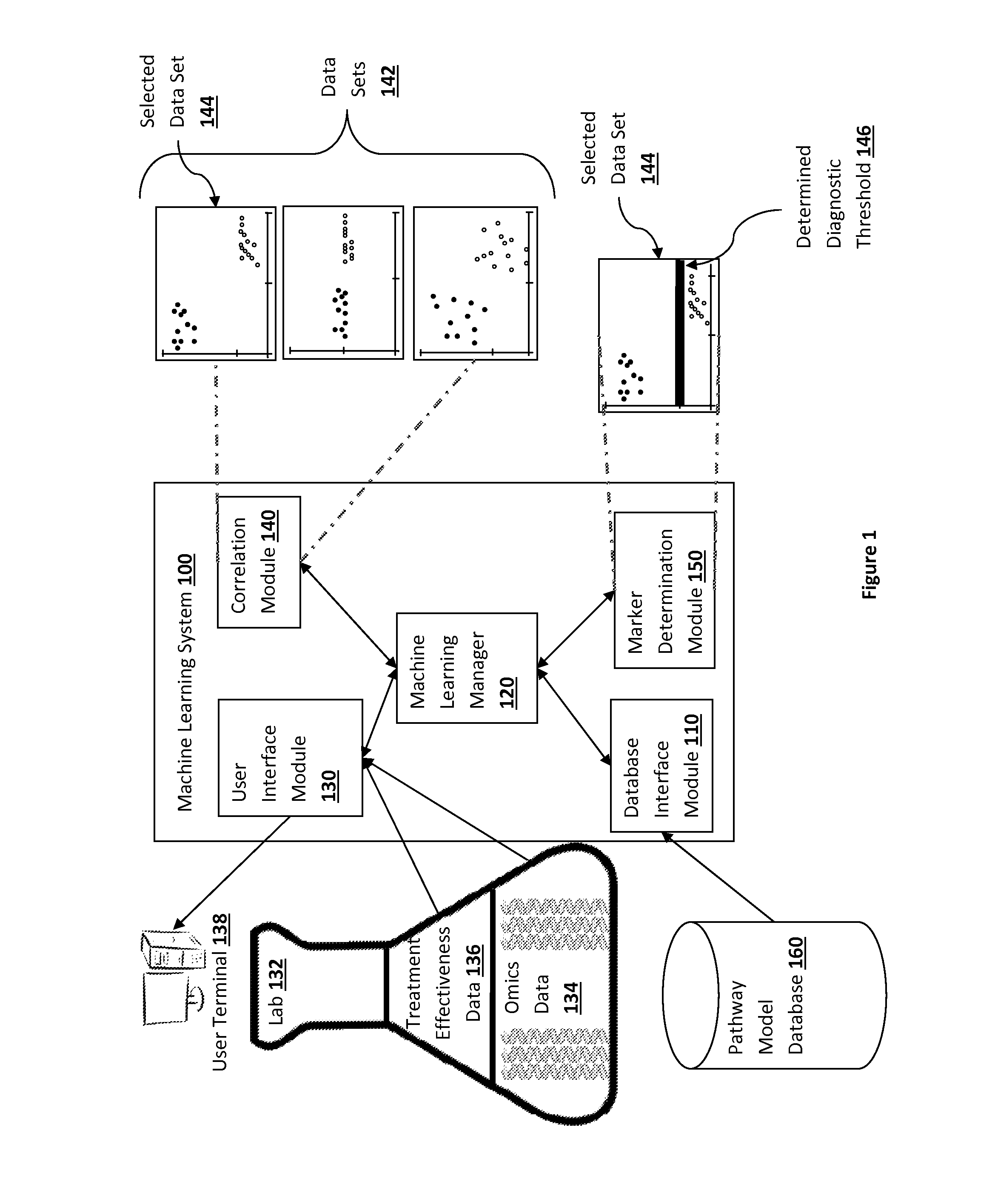
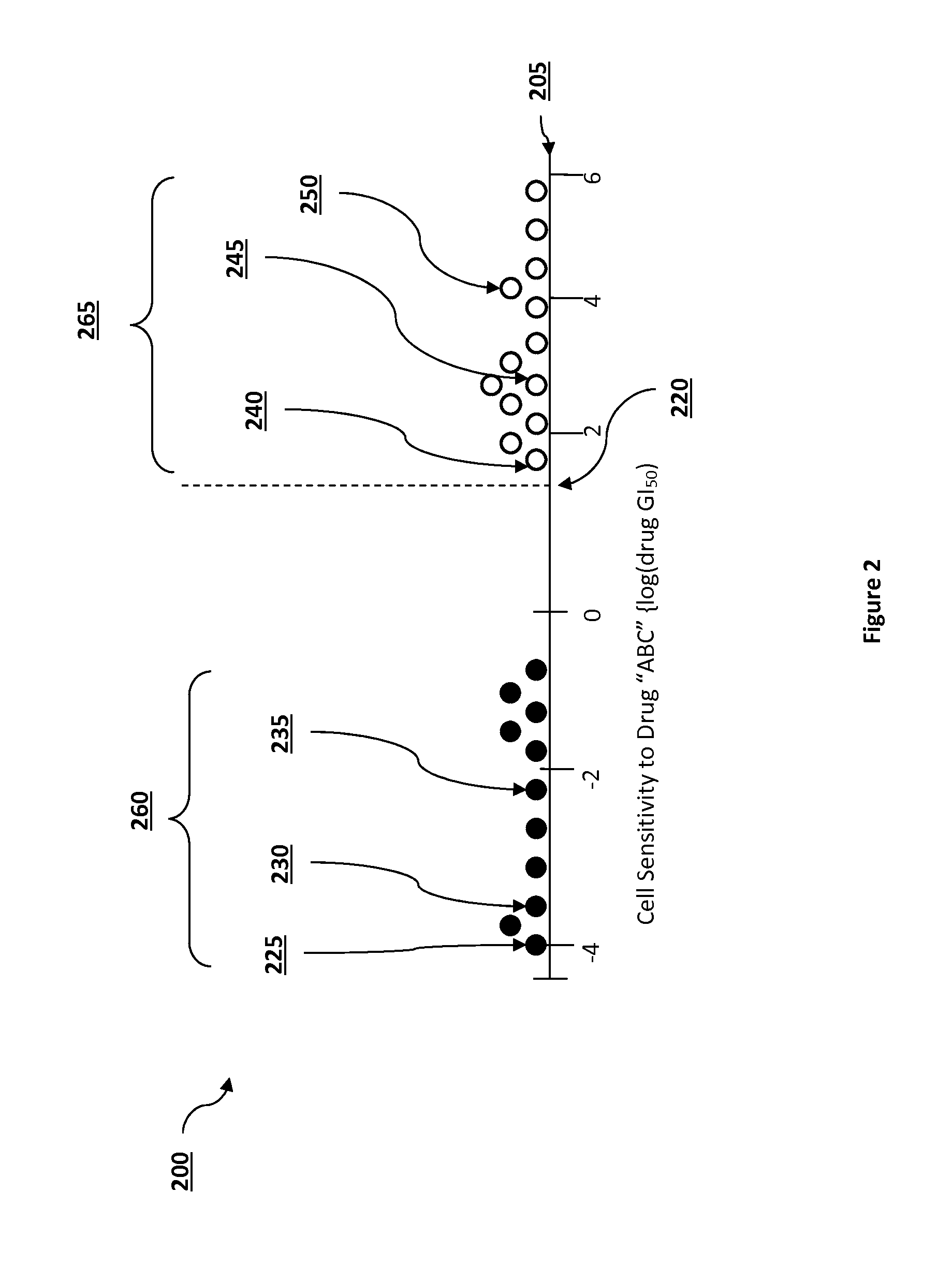
Pathway analysis for identification of diagnostic tests
ActiveUS20150006445A1Expected effectivenessEasy to collectOrganic active ingredientsOrganic chemistryDiagnostic testPathway analysis
The present inventive subject matter provides apparatus, systems, and methods in which a diagnostic test is identified, where the diagnostic test is for determining whether a particular treatment is effective for a particular patient based on one or more characteristics of a patient's cells. When a treatment is developed with the potential to treat one or more diseases, the drug can have different effects on different cell lines related to the diseases. A machine learning system is programmed to infer a measurable cell characteristic, out of many different measurable cell characteristics, that has a desirable correlation with the sensitivity data of different cell lines to a treatment. The machine learning system is programmed to then determine, based on the correlation, a threshold level of the cell characteristic the patient should exhibit in order to recommend administering the treatment.
Owner:NANTOMICS LLC +1
Educational program assessment using curriculum progression pathway analysis
ActiveUS8666300B2Simple methodElectrical appliancesSpecial data processing applicationsPathway analysisPerformance index
A curriculum progression pathway analysis assesses an educational program having multiple activities defining multiple learner tracks by organizing the learners into groups based on the learner tracks and, for each learner group, calculating a baseline average score, a post-activity average score, and statistical differences between the baseline average score and the post-activity average scores. Any significant statistical differences are identified, and a report may be generated describing statistical conclusions and inferences. The invention can further provide a domain-based learner analysis for curriculum, a domain-based learner analysis for activities, and a learner retention analysis. In an application for continuing medical education the performance index test includes a clinical vignette, a set of statements which are moved to either a best practices column or a not best practices column, and an indicator for an allowable number of moves to achieve a perfect score.
Owner:REALCME
Method for analyzing iTRAQ (isobaric Tags for Relative and Absolute Quantitation) data
InactiveCN102321733AMicrobiological testing/measurementBiological testingPathway analysisAnalysis data
The invention designs a method for analyzing iTRAQ (isobaric Tags for Relative and Absolute Quantitation) data aiming at the characteristics of iTRAQ protein quantitative data analysis. The method has a main flow which comprises the following steps of: 1, screening differential protein: screening the initial iTRAQ data in groups to acquire a difference result; 2, performing GO (Gene Ontology) analysis: performing query mapping on a GO database to acquire completer gene function information so as to display a directed acyclic graph; 3, performing Pathway analysis which is similar to GO analysis: performing comparative query with a KEGG (Kyoto Encyclopedia of Genes and Genomes) database to construct an interaction pathway between genes; and step 4, performing Gene network analysis: integrating three different interaction relations to obtain a gene interaction network graph.
Owner:SHANGHAI CLUSTER BIOTECH
Method for data analysis of miRNA in animals with reference data based on miRBase database
The invention discloses a method for data analysis of miRNA in animals with reference data based on a miRBase database. The method is characterized by comprising the following steps of preparation, filtration of logout data, genome alignment, small RNA annotation, miRNA feature analysis, miRNA expression quantity analysis, miRNA function and pathway analyses, and result sorting. The method disclosed by the invention has the beneficial effects that an analysis result is complete, and comprises involved miRNA analysis content and other detected small RNA information annotation, automated analysis is achieved, and statistics and sorting of all analysis content and results can be automatically and rapidly conducted only by providing related database information. All operation steps are visible, and thus errors can be conveniently checked, applied command lines and parameters and log results generated in operation can all be recorded when analysis is conducted step by step, and once a program fails during running, the errors can be rapidly queried.
Owner:南京派森诺基因科技有限公司
System, method and computer product for predicting biological pathways
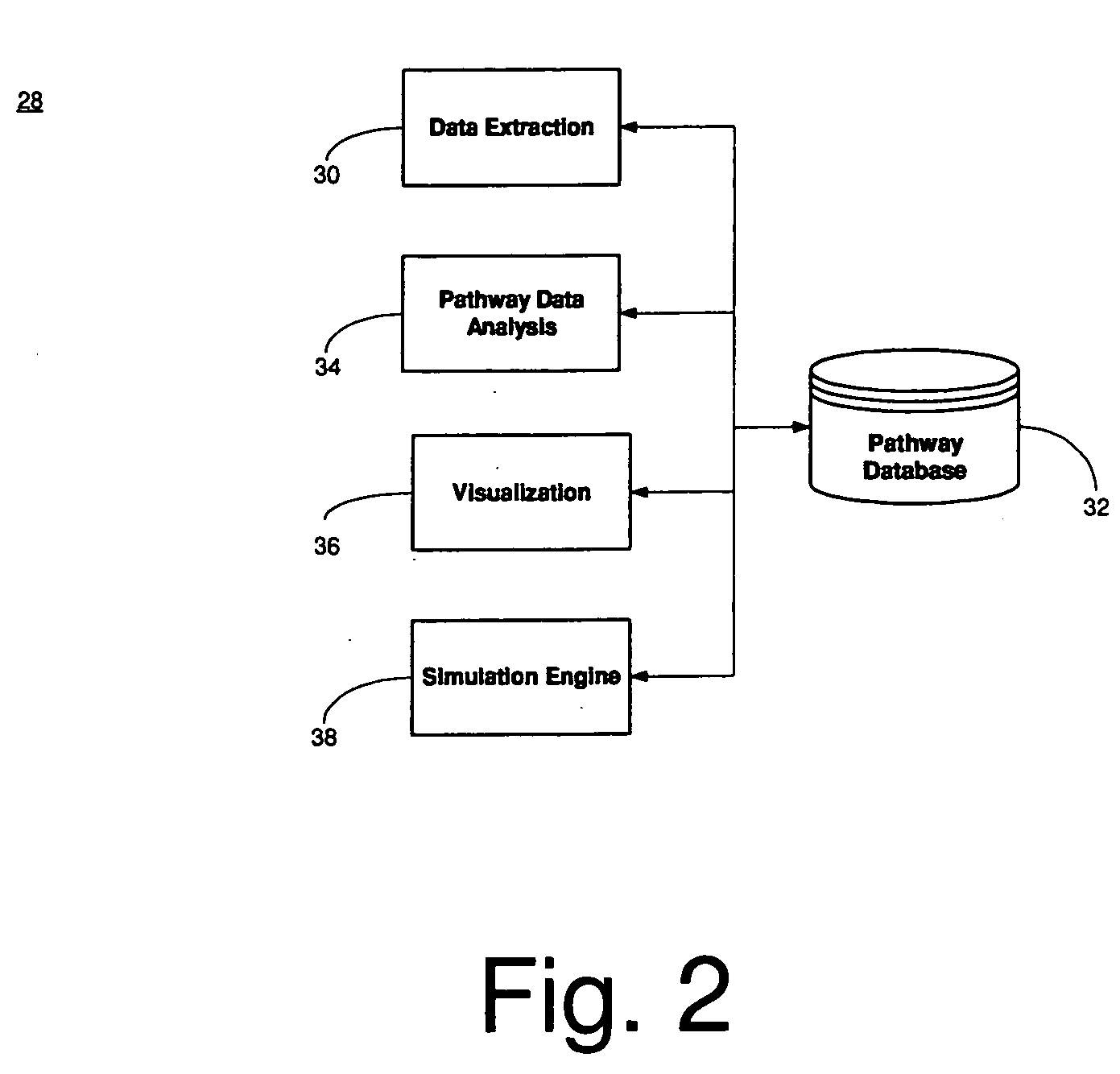
InactiveUS20050004785A1Data visualisationAnalogue computers for chemical processesData ingestionPathway analysis
System, method and computer product for predicting biological pathways. In this disclosure, a data extraction module automatically extracts biological data from biological data sources. A pathway database contains the extracted biological data. A pathway analysis module assimilates the biological data into a hypotheses prediction for generating a pathway. A visualization module generates a visual representation of the pathway generated by the pathway analysis module.
Owner:GENERAL ELECTRIC CO
Method for data analysis of miRNA in plants with reference data based on miRBase database
The invention discloses a method for data analysis of miRNA in plants with reference data based on a miRBase database. The method is characterized by comprising the following steps of filtration of logout data, sRNA classification and annotation, miRNA differential analysis, miRNA function and pathway analyses, miRNA sequence feature analysis, and result sorting. The method disclosed by the invention has the beneficial effects that the proper analysis method is adopted aiming at the features of the miRNA of the plants; a result is comprehensive and comprises involved miRNA analysis content andother detected small RNA information annotation; all analysis results are automatically sorted; after analysis of each part is completed, automatic statistics of the results is achieved, and the results are visualized, classified and sorted, so that result arrangement is clear and is directly used for report generation; all operation steps are visible, and errors can be conveniently queried.
Owner:南京派森诺基因科技有限公司
Mass spectrum data missing value filling method and system based on non-negative matrix factorization
ActiveCN111859275AGood filling accuracyImprove robustnessComplex mathematical operationsData setPathway analysis
The invention relates to a mass spectrum data missing value filling method and system based on non-negative matrix factorization, and the method comprises the steps: carrying out the pre-filling of missing values of a data set matrix, and obtaining a missing-free initial data matrix; carrying out logarithm transformation on all elements in the initial data matrix without loss; taking a group of dimension parameters of non-negative matrix factorization, and respectively carrying out non-negative matrix factorization to obtain a group of corresponding reconstruction matrixes; performing exponential transformation on the element values of the reconstructed matrix; calculating reconstruction errors between all reconstruction matrixes after exponential transformation and the missing-free initial data matrix; calculating corresponding weights under different reconstruction matrixes according to the reconstruction errors; performing weighted average on the reconstruction matrix to obtain a weighted reconstruction matrix; filling the missing positions in the data set matrix with the element values at the corresponding positions in the weighted reconstruction matrix; and carrying out characteristic metabolite identification and pathway analysis based on the missing-free final data matrix. According to the method, the data filling precision can be improved.
Owner:XIAMEN UNIV
Laser microdissection and microarray analysis of breast tumors reveal estrogen receptor related genes and pathways
InactiveUS20080305959A1Sugar derivativesMicrobiological testing/measurementPathway analysisEstrogen receptor
About 70% to 80% of breast cancers express estrogen receptor-α (ERα), and estrogens play important roles in the development and growth of hormone-dependent tumors. Together with lymph node metastasis, tumor size and histological grade, ER status is considered one of the prognostic factors in breast cancer, and an indicator for hormonal treatment. 147 genes and 112 genes with significant P-value and having significant differential expression between ER+ and ER− tumors were identified from the LCM data set and bulk tissue data set, respectively. 61 genes were found to be common in both data sets, while 85 genes were unique to the LCM data set and 51 genes were present only in the bulk tumor data set. Pathway analysis with the 85 genes using Gene Ontology suggested that genes involved in endocytosis, ceramide generation, Ras / ERK / Ark cascade, and JAT-STAT pathway may play roles related to ER. The gene profiling with LCM-captured tumor cells provides a unique approach to characterize and study epithelial tumor cells and to gain an insight into signaling pathways associated with ER.
Owner:VERIDEX LCC
Pathway analysis for providing predictive information
A method for assigning ranking scores to pathways in a set of pathways for classifying patients is disclosed. The method comprises the steps of comparing biomolecular datasets from different groups of patients and performing an analysis in order to assign ranking scores to pathways in a set of pathways. Furthermore, a method for using cancer pathway evaluation to support clinical decision making is disclosed. This assessment is further used for stratifying ovarian cancer patients based on chemosensitivity to platinum based drugs, the standard chemotherapy. We present the method for evaluation and ranking of the most relevant pathways responsible for platinum sensitivity. Clinical decision support software system should be able to then visualize this information for a clinician, contextualize it within a patient data set and help make a final decision on the potential responsiveness.
Owner:KONINKLIJKE PHILIPS ELECTRONICS NV
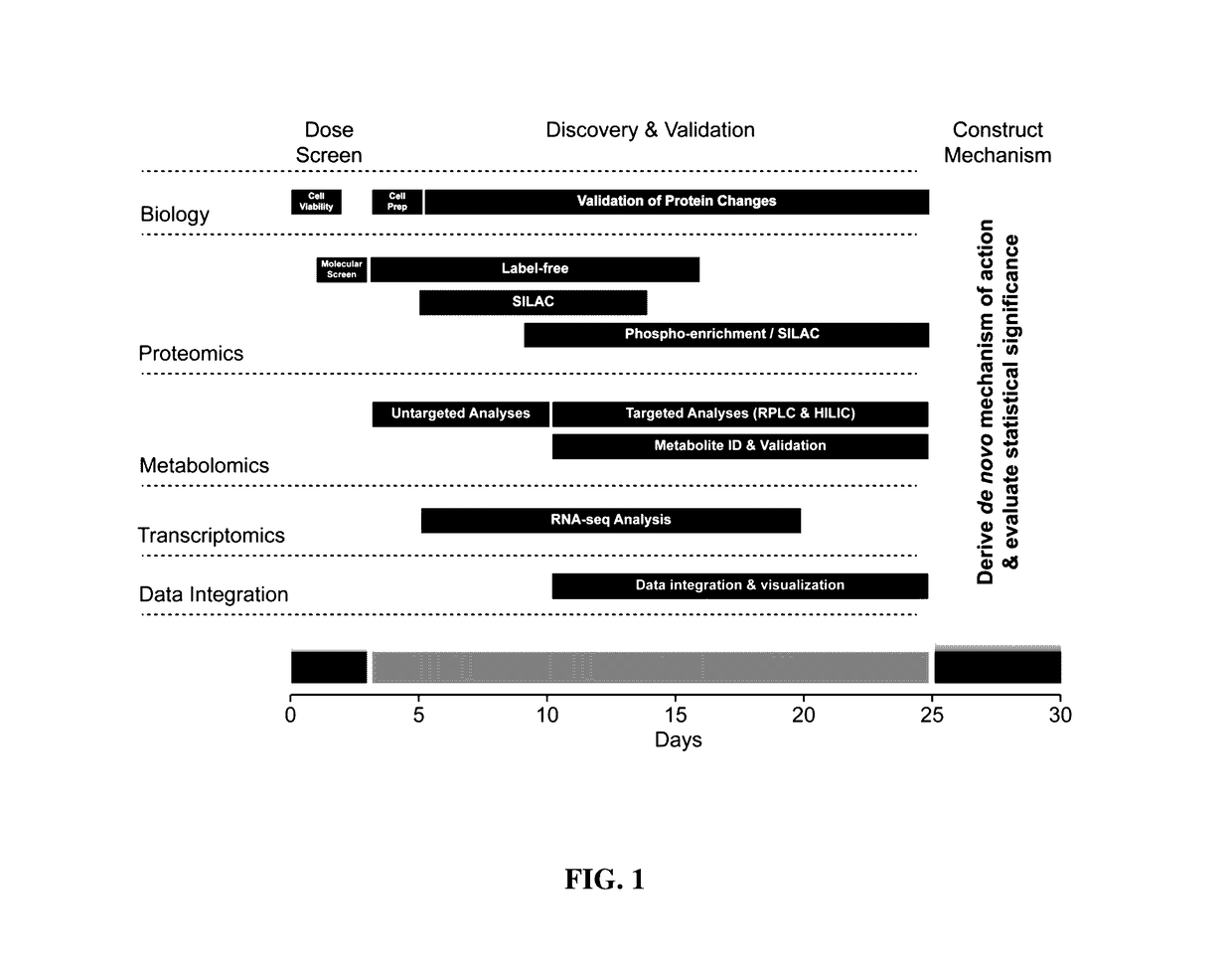
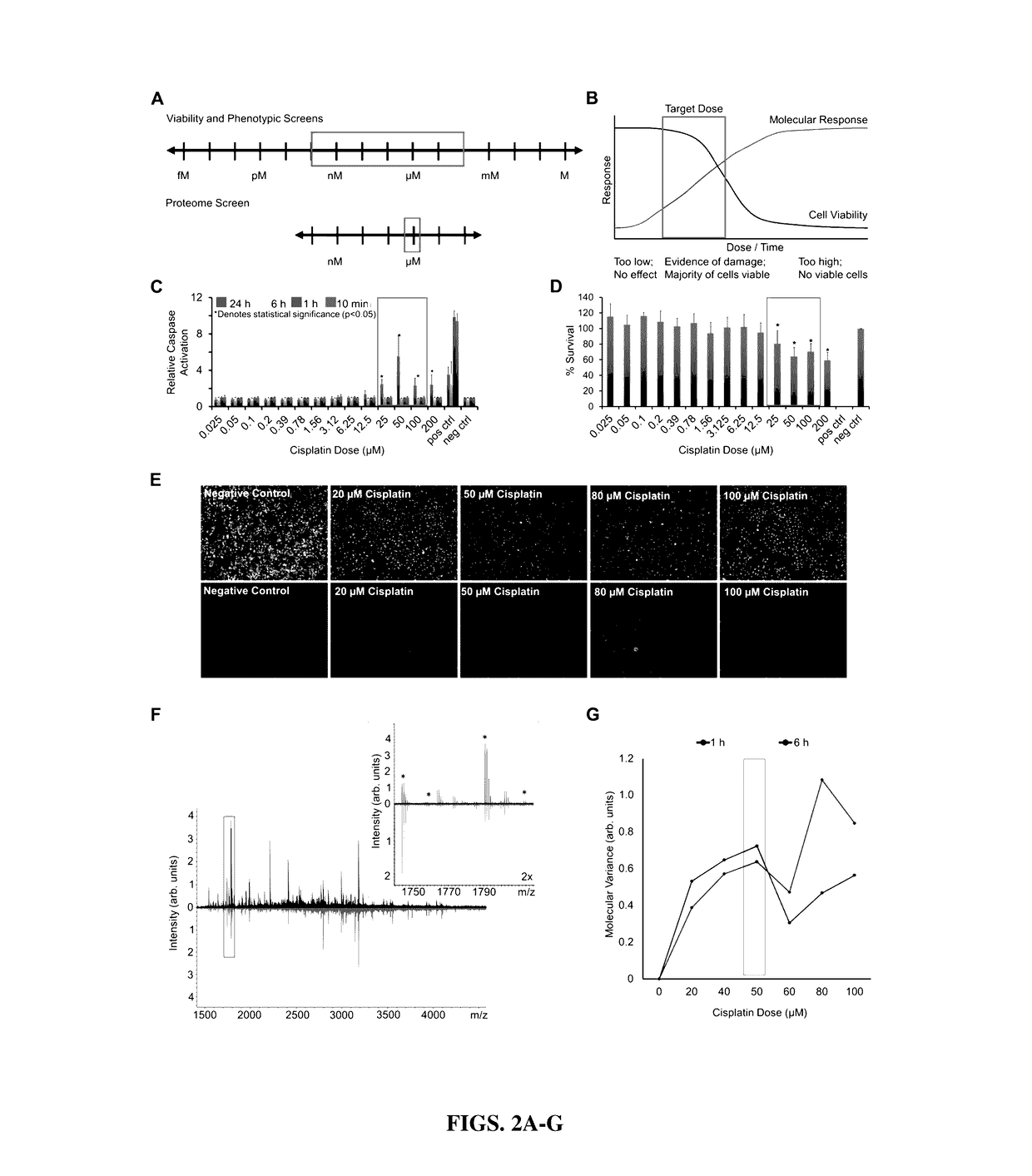
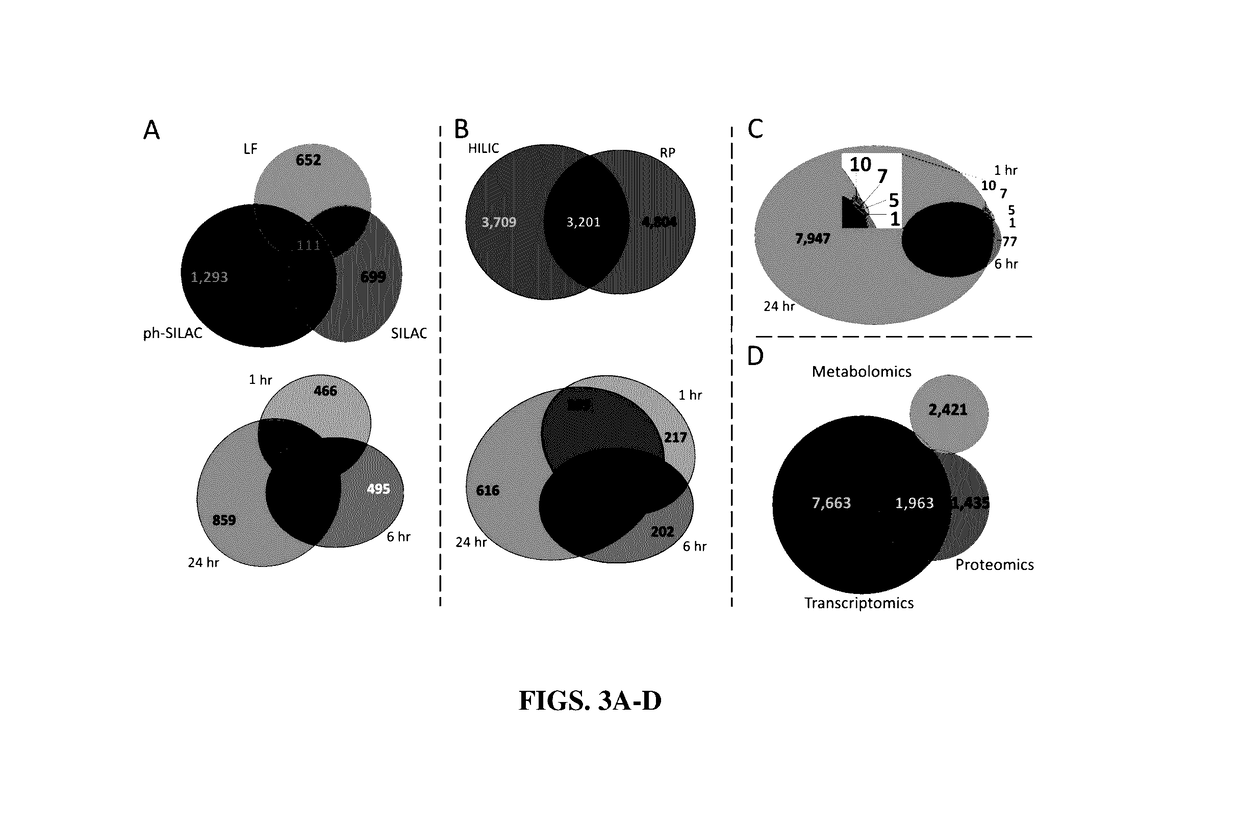
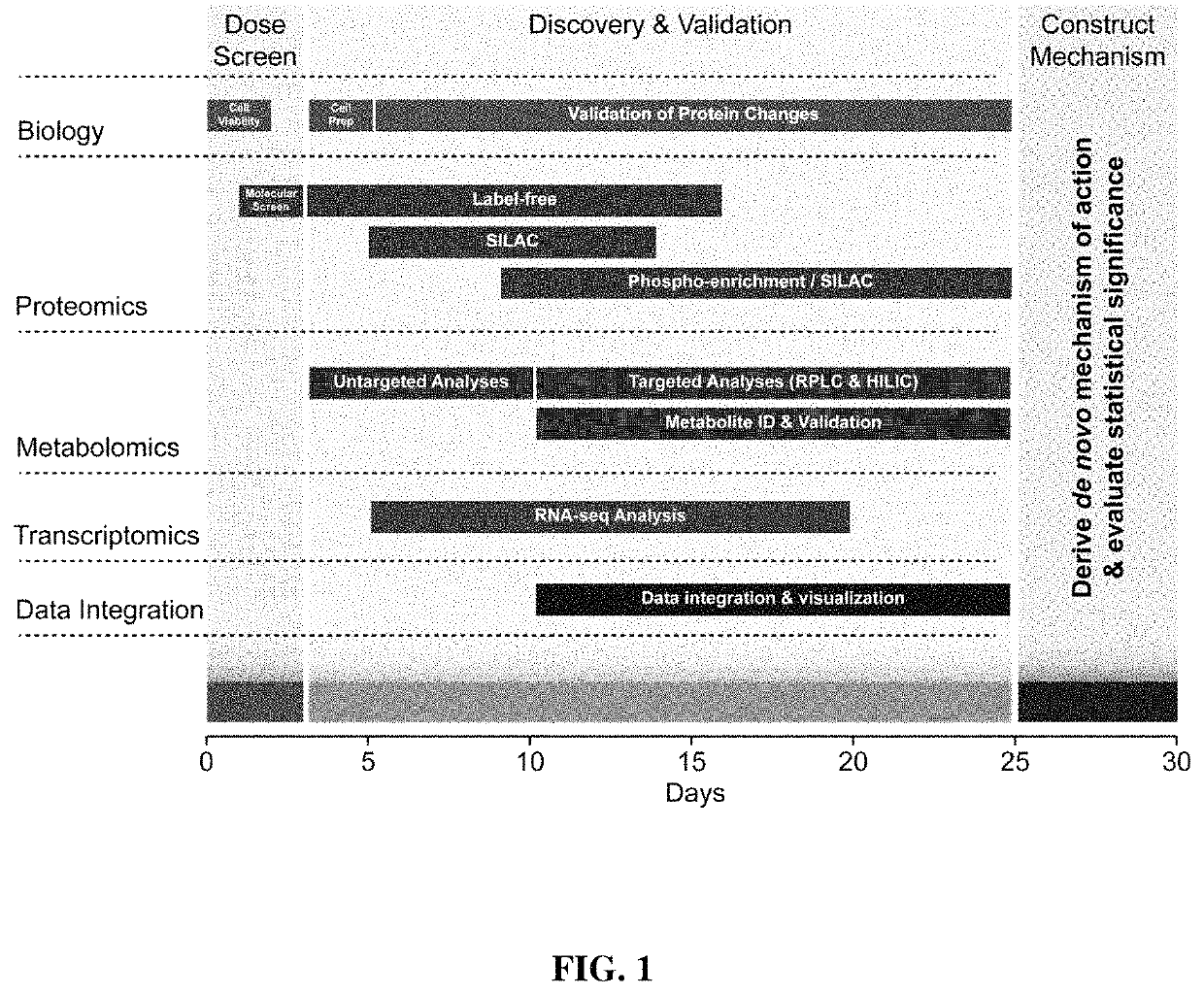
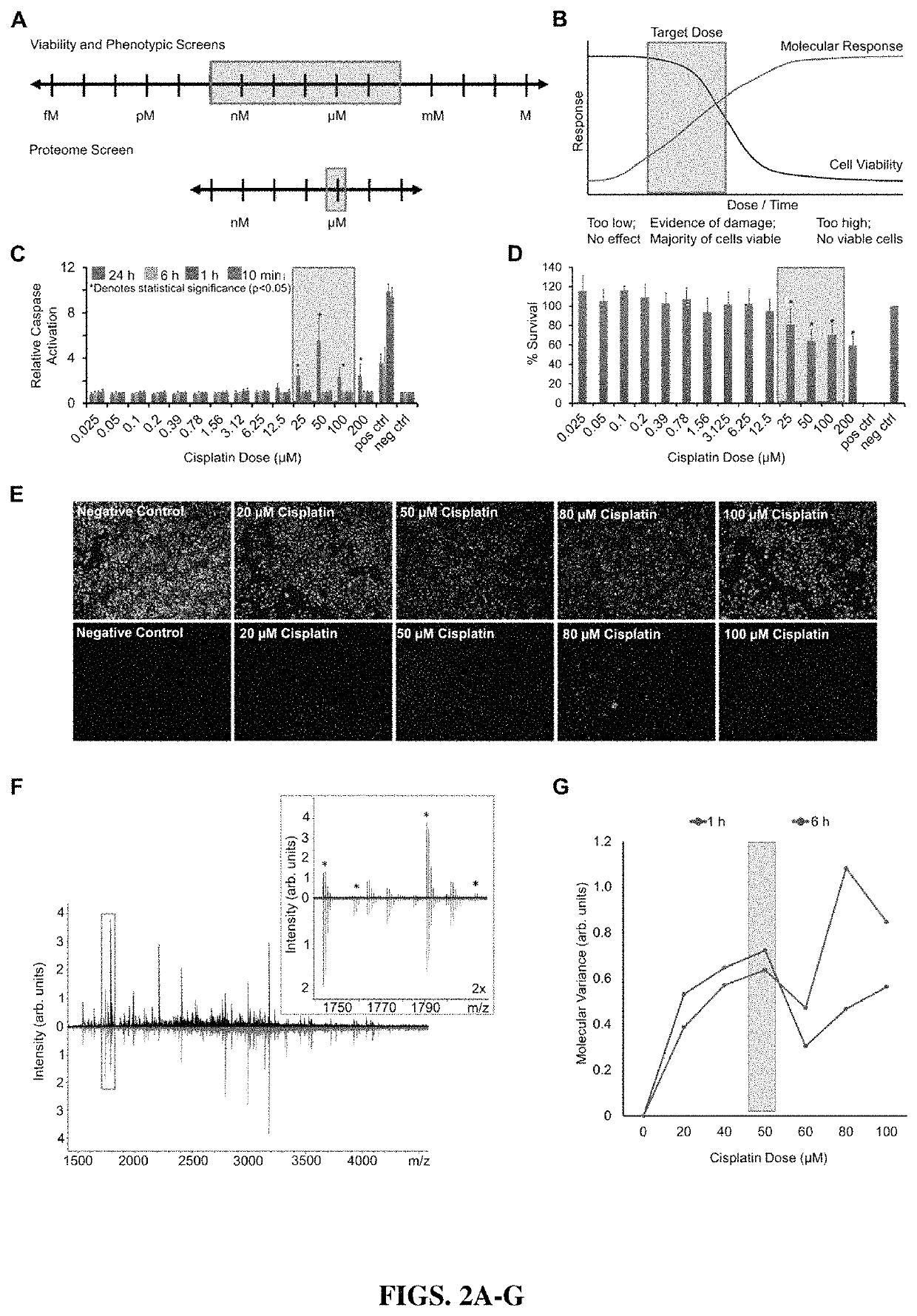
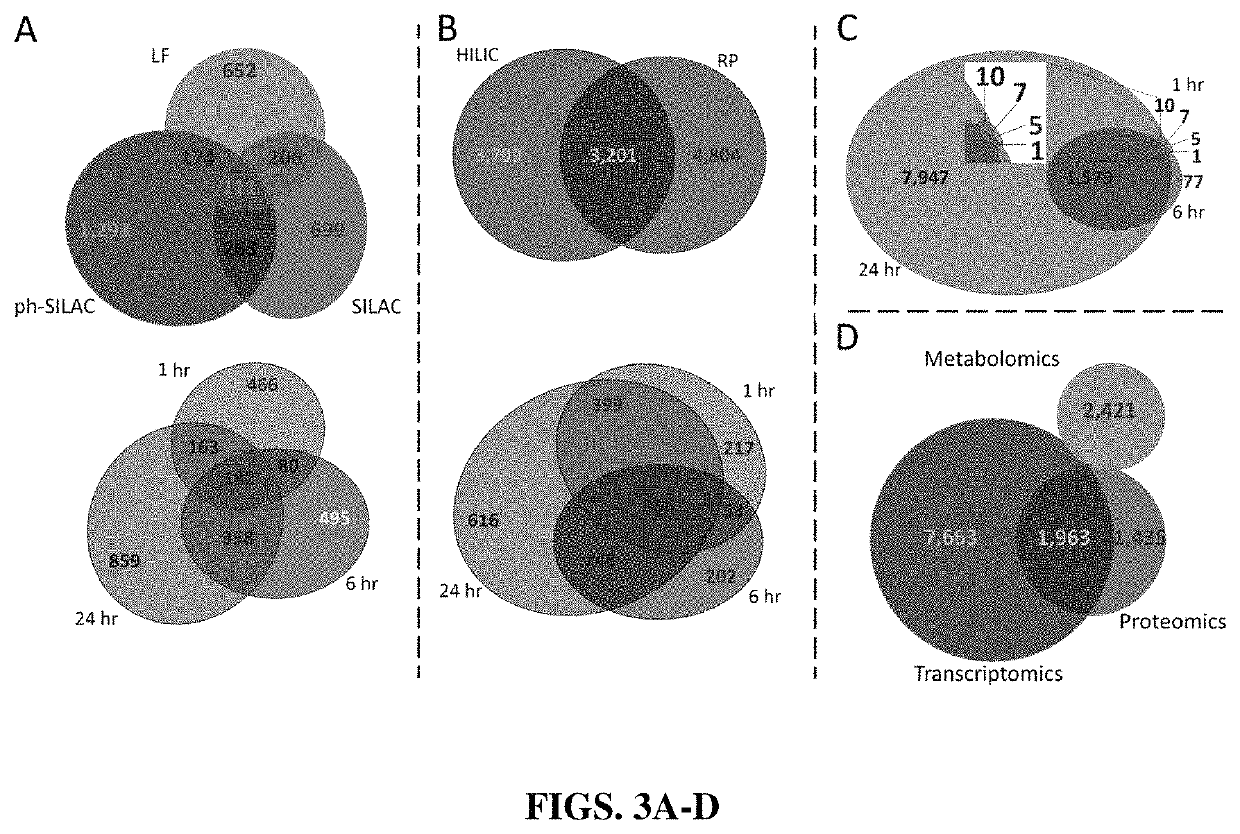
High-throughput, multi-omics approach to determine and validate de novo global mechanisms of action for drugs and toxins
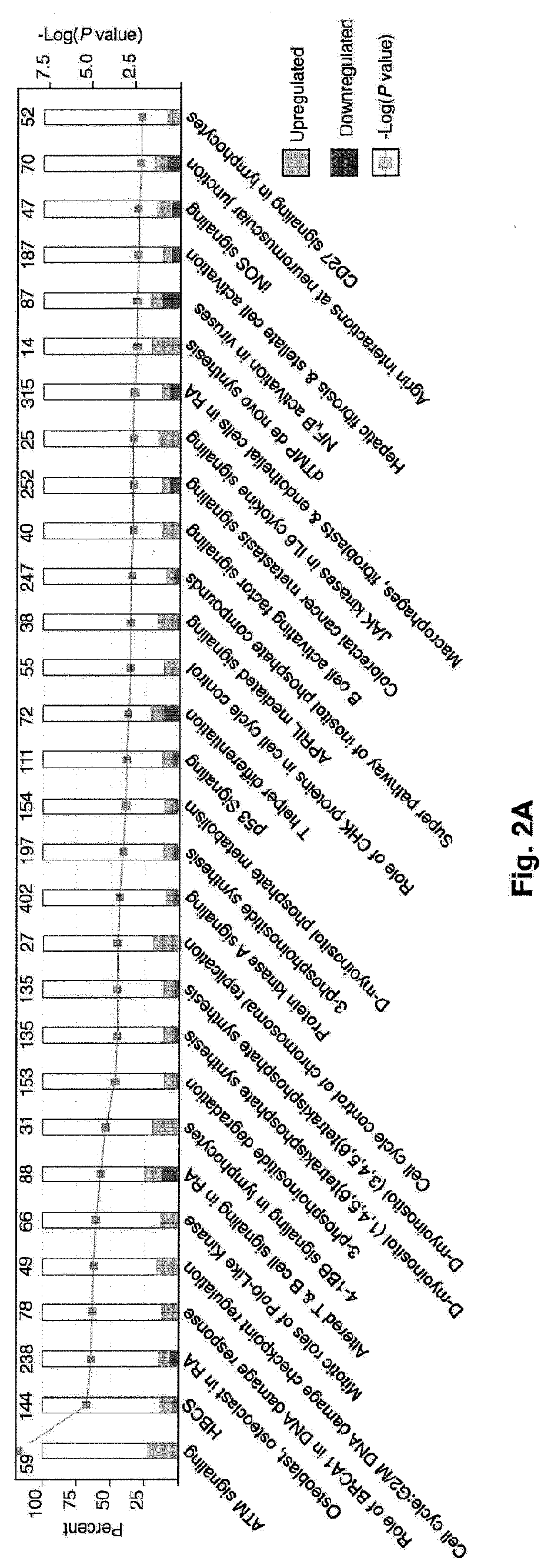
The present disclosure provides for rapid identification of mechanism of action (MOA) for drugs and toxins, and does so in a rapid (30 days or less) fashion. The methods use a combination of high throughput bioinformatics and pathway analysis that examine a wide variety of biological parametics.
Owner:VANDERBILT UNIV
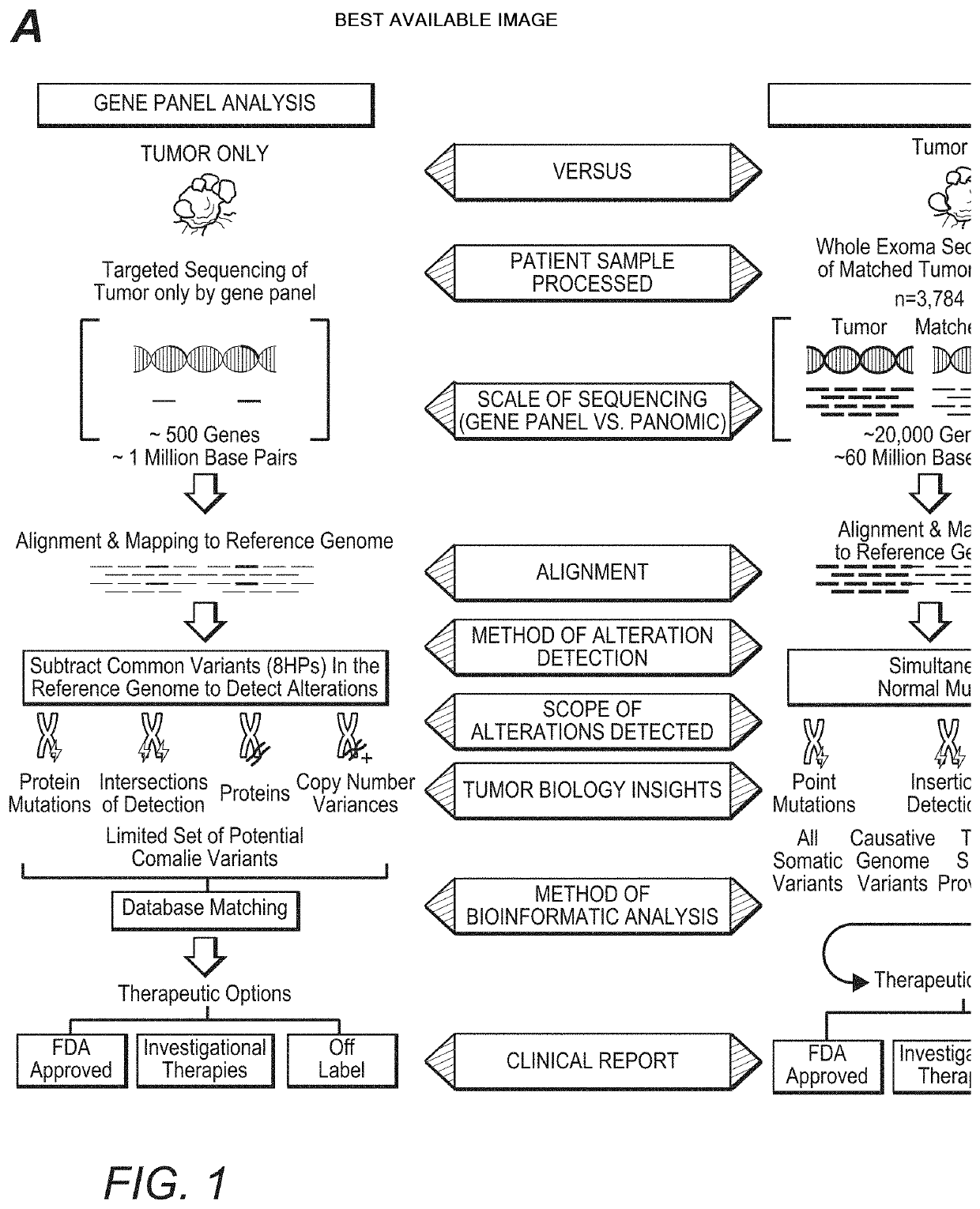
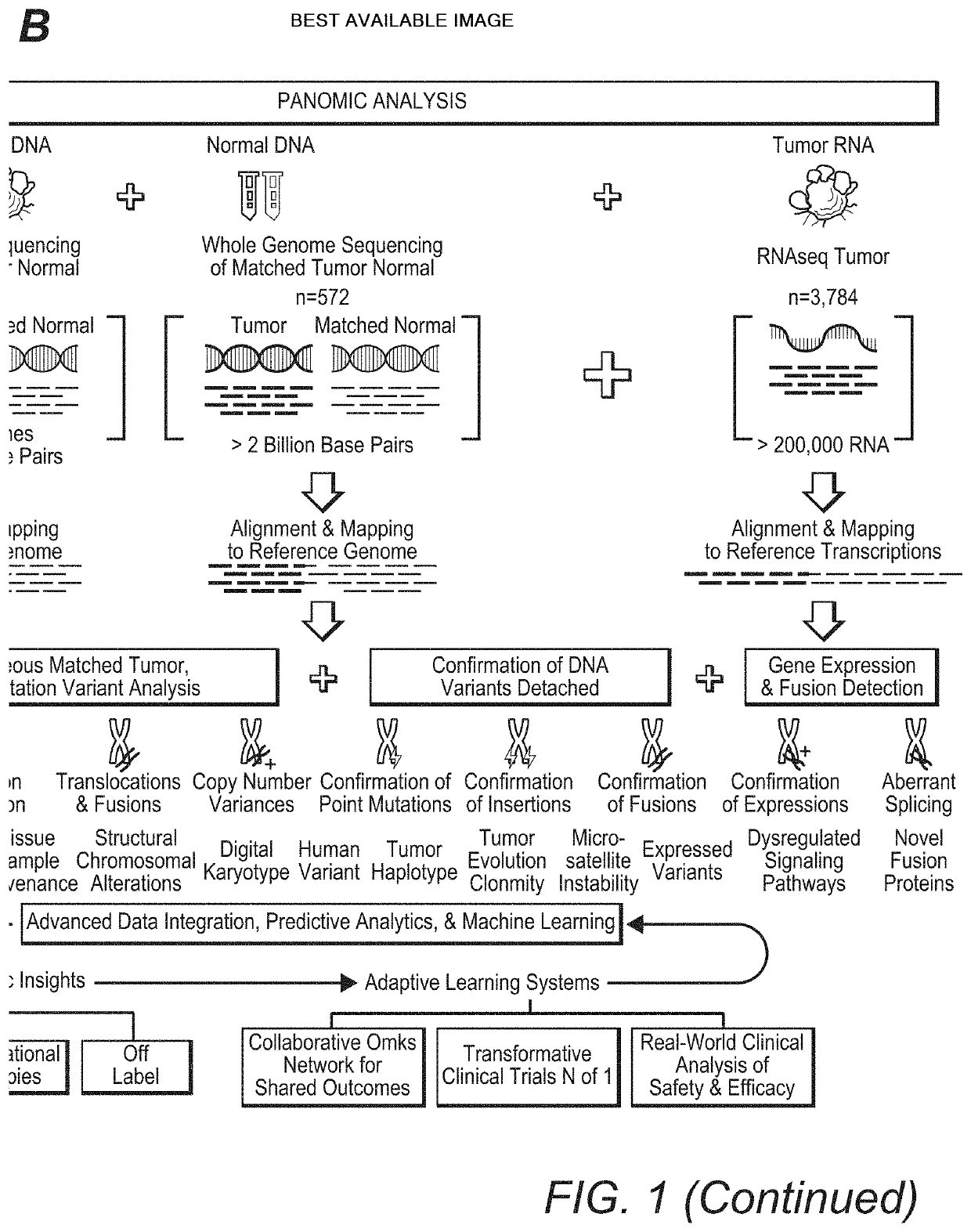
High Throughput Patient Genomic Sequencing and Clinical Reporting Systems
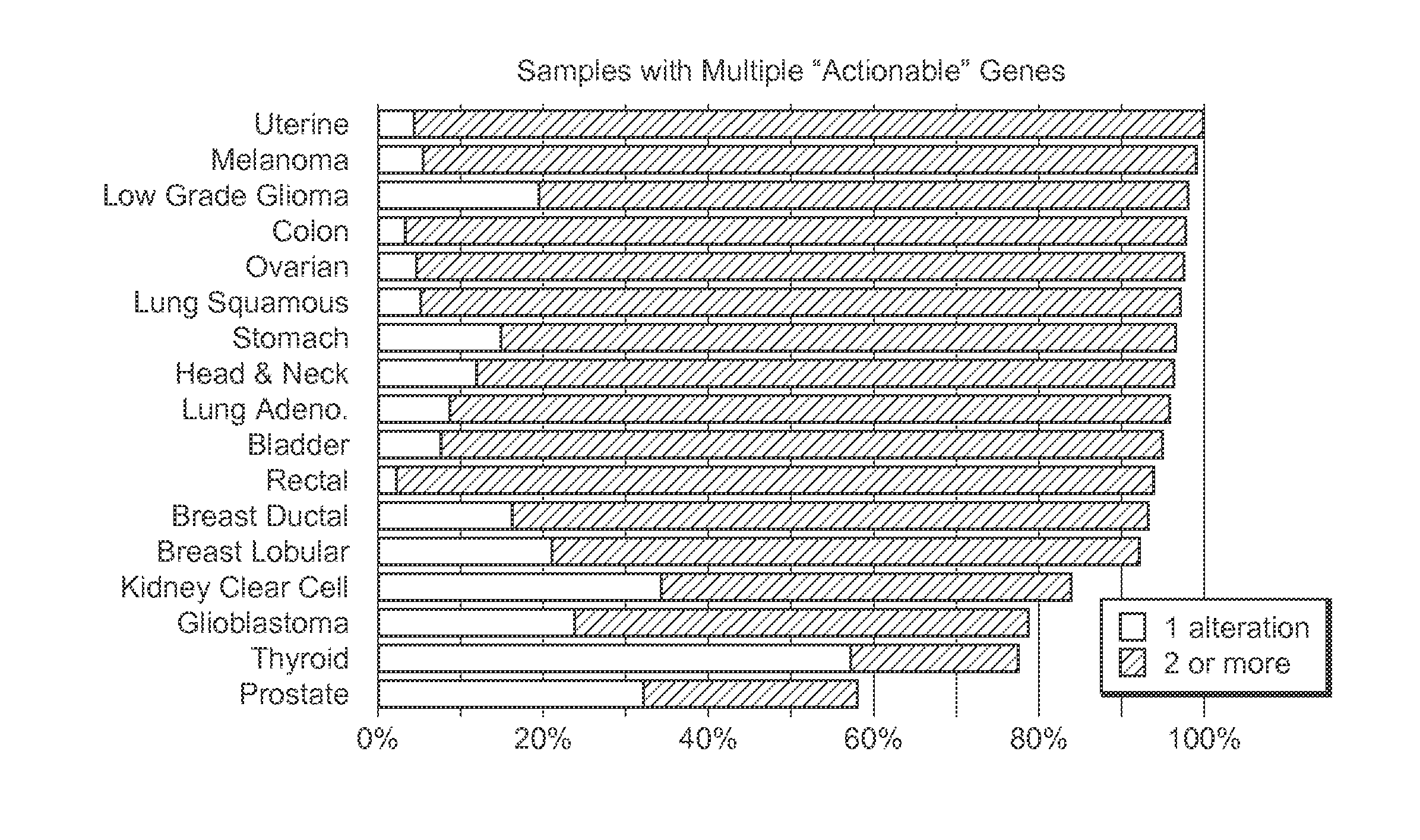
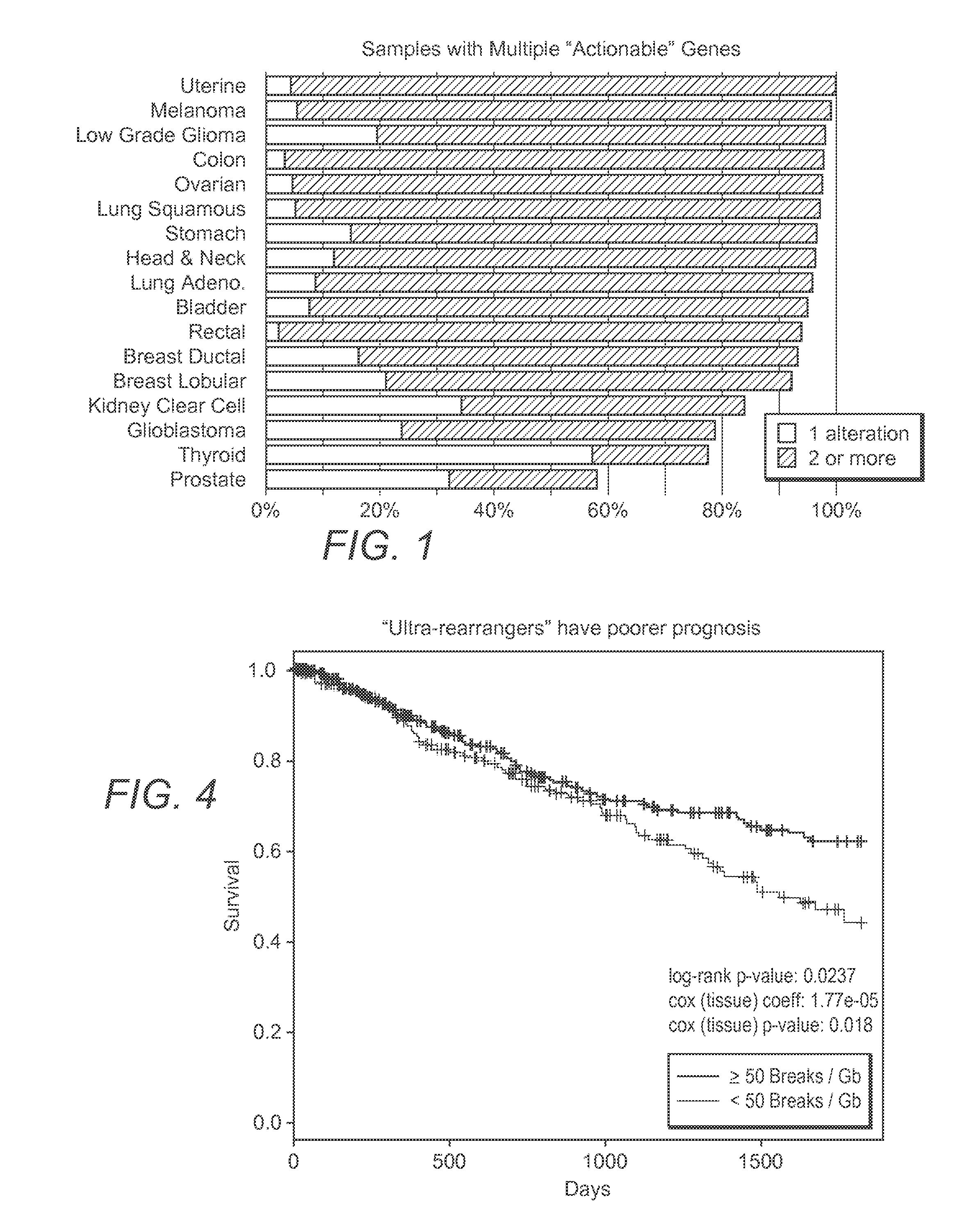
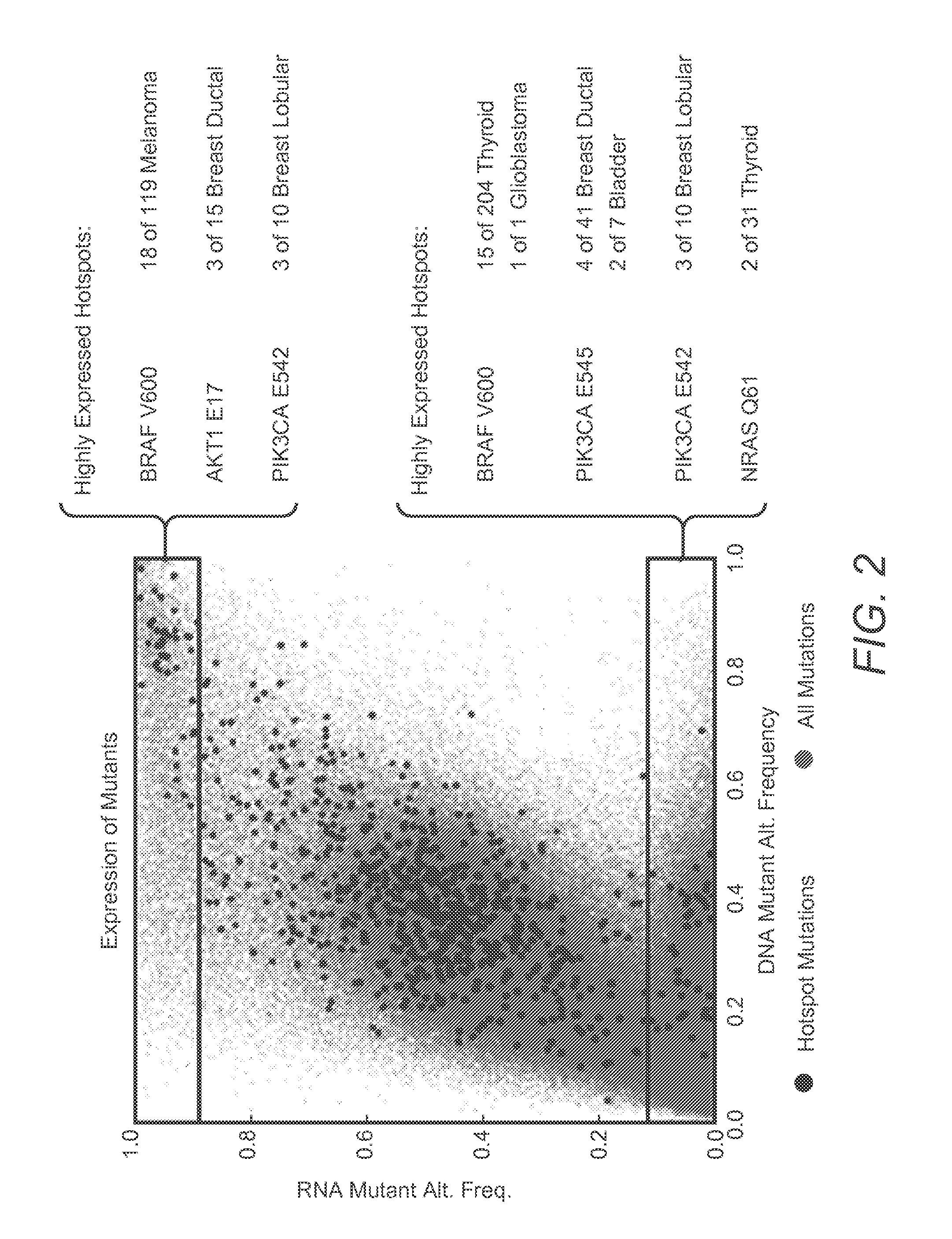
InactiveUS20200273537A1Accurate predictionEasy to identifyProteomicsGenomicsGenomic sequencingPathway analysis
Contemplated panomic systems and methods significantly improve accuracy of genetic testing by taking into account matched normal data and expression levels of various genes in diseased tissue. Analysis and physician guidance is further improved by combining so identified clinically relevant changes with pathway analysis to thereby allow for classification of a tumor and / or identification of potentially druggable targets within affected pathways.
Owner:NANTOMICS LLC
Abietane type tricyclic diterpenoid C-14 site hydroxylase
PendingCN114480312AReduced gene expressionReduce expressionFungiMicroorganism based processesPathway analysisCytochrome P450
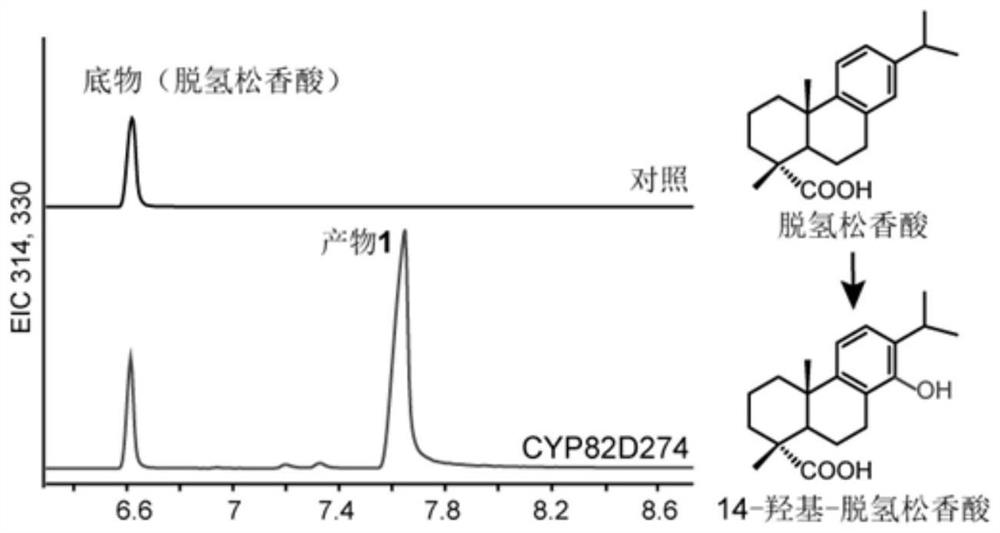
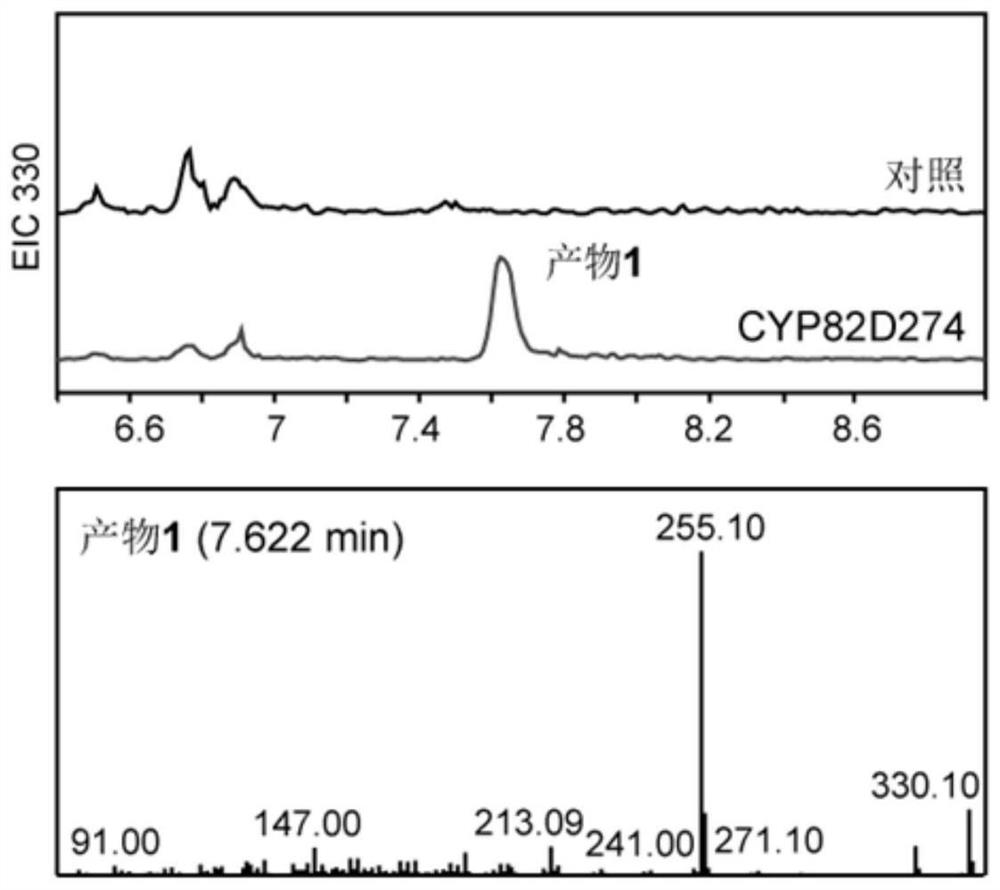
The invention relates to an enzyme for catalyzing hydroxylation of an abietane type tricyclic diterpenoid compound C-14, the enzyme is tripterygium wilfordii cytochrome p450 oxidase CYP82D274, and the invention also relates to polynucleotide CYP82D274 for coding the enzyme, and the polynucleotide CYP82D274 is used for catalyzing hydroxylation of the abietane type tricyclic diterpenoid compound C-14. An enzyme catalysis experiment proves that the triptolide gene has the function of catalyzing hydroxylation of triptolide biosynthesis intermediates, namely miltidiene, dehydroabietic acid and triptobenzene terpene D. The triptolide gene has the advantages that triptolide biosynthesis pathway analysis is further promoted, an important gene element is provided for synthesis biological production of the triptolide intermediates, and the triptolide gene has a wide application prospect. The method has important significance on synthesis regulation and control of abietane type tricyclic diterpenoid compounds such as triptolide and the like.
Owner:BEIJING SHIJITAN HOSPITAL CAPITAL MEDICAL UNIVERSTY
Metabonomics research method of Xinshao prescription on cerebral ischemia-reperfusion injury based on gc-ms combination technology
The invention provides a GC-MS (Gas Chromatography-Mass Spectrometer) combined technology-based metabonomics research method for resistance of an asarum and red peony root composition to a cerebral ischemia-reperfusion injury. The method is realized through the following steps: (1) detecting and analyzing a serum sample and a urine sample of a mouse by adopting a GC-MS combined technology; (2) analyzing data by using PCA (Primary Component Analysis) and PLS-DA (Partial Least Square)-DA (Discriminant Analysis),and performing metabolite identification in combination with SPSS17.0 by using an NIST (National Institute of Standards and Technology) database to find out a final differential metabolite; (3) performing pathway analysis on the differential metabolite through MetPA online analysis software. According to the method, by analysis of the serum sample and the urine sample of the mouse through the GC-MS combined technology, respective identification of the differential metabolites of the serum sample and the urine sample of the mouse, and metabolic pathway analysis of the differential metabolites, the action mechanism of the asarum and red peony root composition resisting the cerebral ischemia-reperfusion injury can be comprehensively evaluated from the overall level, so that a demonstrative research is provided for explanation of the metabonomics and the action mechanism of a traditional Chinese medicine composition.
Owner:GUIZHOU MEDICAL UNIV
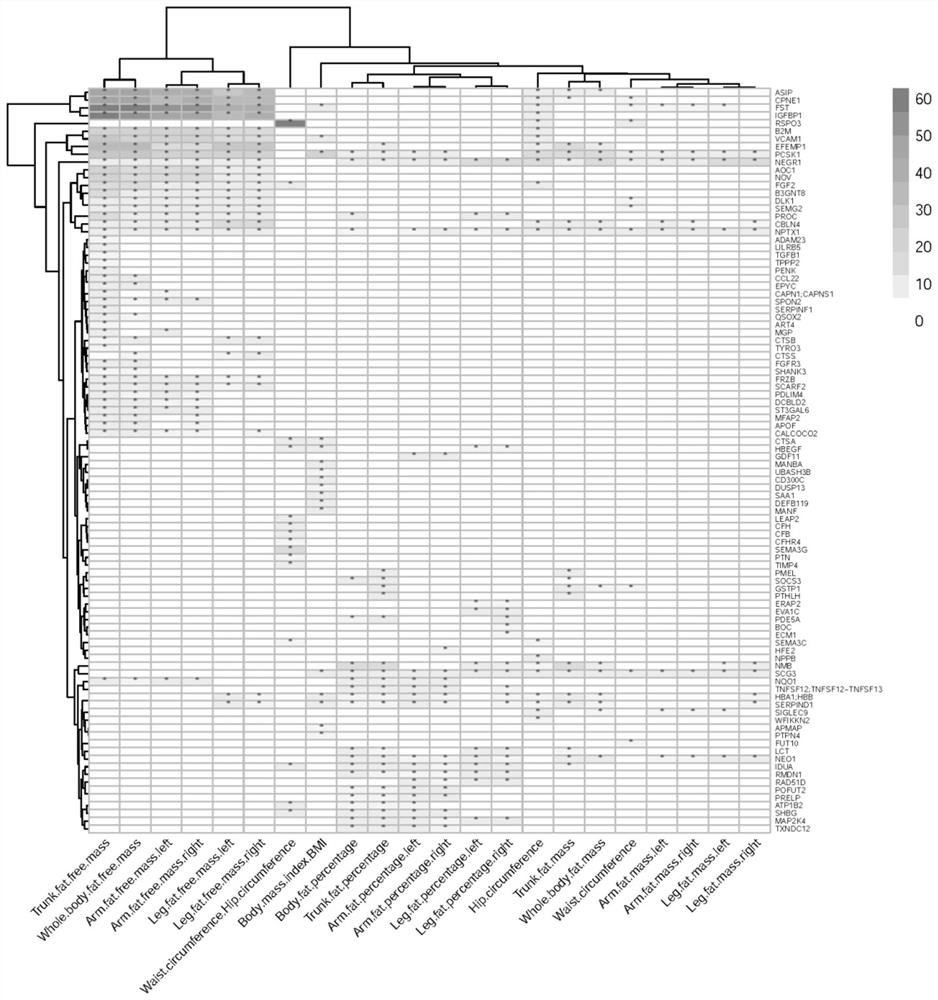
Method for integrating and analyzing plasma proteome, genome and obesity related traits
PendingCN114188029AInhibition biasReliable relationshipHealth-index calculationProteomicsPathway analysisNucleotide
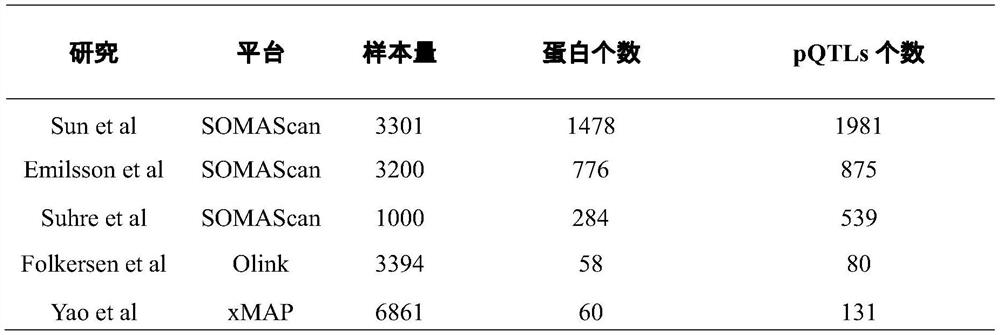
The invention discloses a method for integrating and analyzing plasma proteome, genome and obesity-related traits. The method comprises the following steps: S1, obtaining whole genome associated summary data of the plasma proteome and whole genome associated summary data of the obesity-related traits; s2, screening out SNPs (Single Nucleotide Polymorphisms) which are in significant correlation with the plasma protein in a whole genome range; s3, screening the SNPs which are in significant correlation to obtain independent SNPs, and removing horizontal multiple effects from the independent SNPs; s4, performing double-sample Mendel randomization analysis, and inferring through causal association to obtain plasma protein having causal association with obesity-related traits; s5, performing system clustering analysis to evaluate a structure and a mode of significant causal association; and S6, carrying out gene enrichment analysis and pathway analysis to explore the functional relevance of obesity-related proteins. According to the method, the plasma protein which is causally associated with the obesity-related traits can be obtained, and the determined positive correlation or negative correlation between the plasma protein and the obesity-related traits can be obtained.
Owner:SUZHOU UNIV
High-throughput, multi-omics approach to determine and validate de novo global mechanisms of action for drugs and toxins
The present disclosure provides for rapid identification of mechanism of action (MOA) for drugs and toxins, and does so in a rapid (30 days or less) fashion. The methods use a combination of high throughput bioinformatics and pathway analysis that examine a wide variety of biological parametics.
Owner:VANDERBILT UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com