Mass spectrum data missing value filling method and system based on non-negative matrix factorization
A non-negative matrix decomposition, mass spectrometry data technology, applied in the field of data missing processing, can solve the problems of poor filling value stability, affecting filling performance, result deviation, etc., to achieve stable performance, excellent filling accuracy, and good filling accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
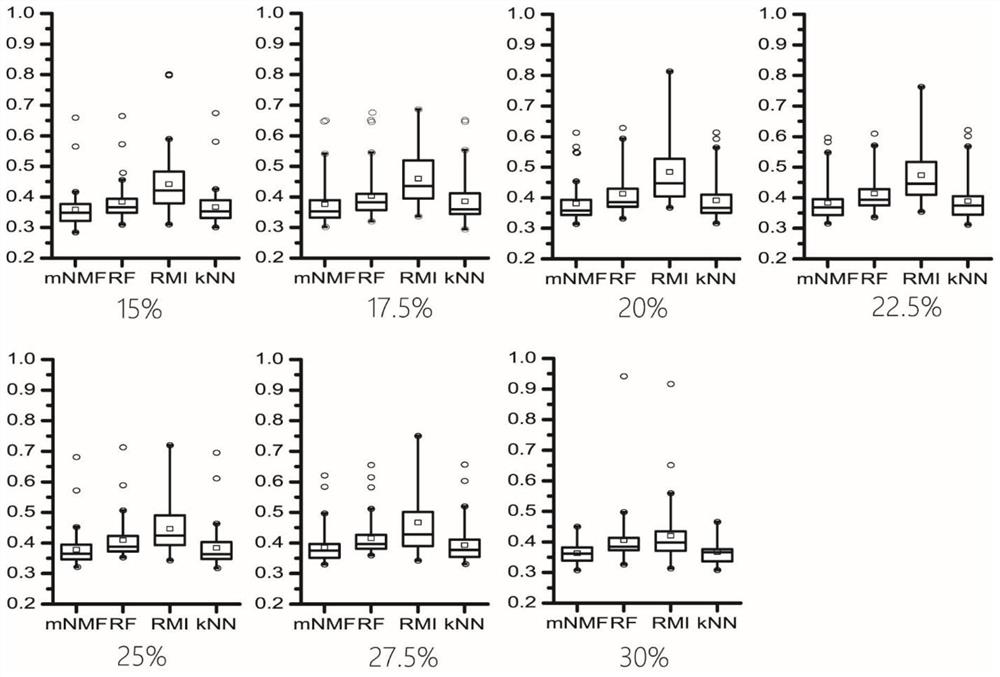
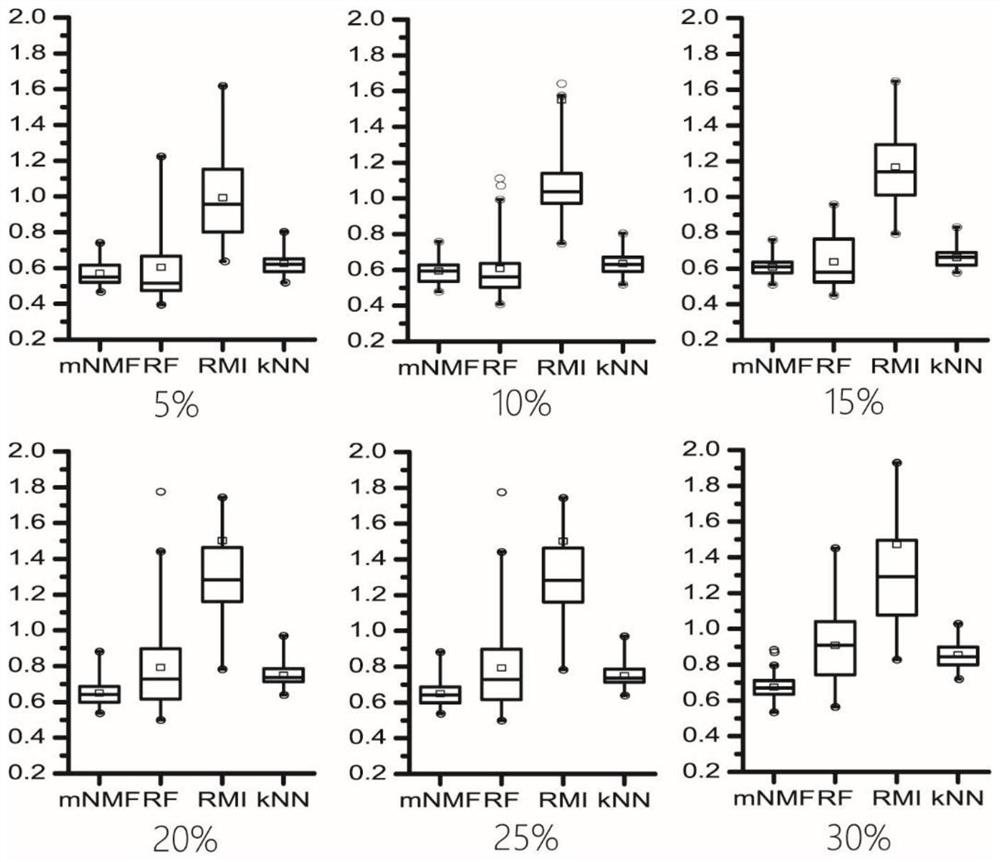
[0092]In this embodiment, the mass spectrometry data sets are respectively placed in the three missing modes of MCAR, MNAR and MM. In each missing mode, different proportions of missing values are simulated, and three disclosed missing value filling methods and the present invention are used. The method performs filling respectively, and the filling accuracy is evaluated through the improved NRMSE, thereby verifying the performance of the method of the present invention.
[0093] 1. Research object
[0094] Collection of mass spectrometry data sets: ①Data set 1 is from the experiment in the literature "Identification of Altered Metabolic Pathways in Plasma and CSF in Mild Cognitive Impairment and Alzheimer's Disease Using Metabolomics" published by Trushina et al. (Trushina, Dutta, Persson, Mielke, & Petersen, 2013). A plasma data set based on liquid chromatography / mass spectrometry (LC-MS) non-targeted metabolomics study of Alzheimer's disease (AD), the data set consists of...
Embodiment 2
[0112] In this example, a mass spectrometry data set is used to generate a data matrix simulating the missing pattern of MM, and extreme values of different proportions are added, followed by filling with three published missing value filling methods and the method of the present invention, and evaluating the filling by the improved NRMSE accuracy, thereby verifying the performance of the method of the present invention.
[0113] 1. Research object
[0114] Mass Spectrometry Dataset Collection: Mass Spectrometry Dataset 1 is from the experiments published by Trushina et al. "Identification of Altered Metabolic Pathways in Plasma and CSF in Mild Cognitive Impairment and Alzheimer's Disease Using Metabolomics", which is based on liquid chromatography / mass spectrometry (LC-MS) The plasma data set of Alzheimer's disease (AD) non-targeted metabolomics research, the data set contains 557 assigned metabolites, 45 samples, and the missing value ratio of the data set is 11.7%;
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com