Method for data analysis of miRNA in animals with reference data based on miRBase database
A data analysis and database technology, applied in genomics, bioinformatics, instrumentation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] The present invention will be further described by the following examples, but these examples should not be used to explain the limitation of the present invention.
[0042] see figure 1 , the detailed steps of the present invention include:
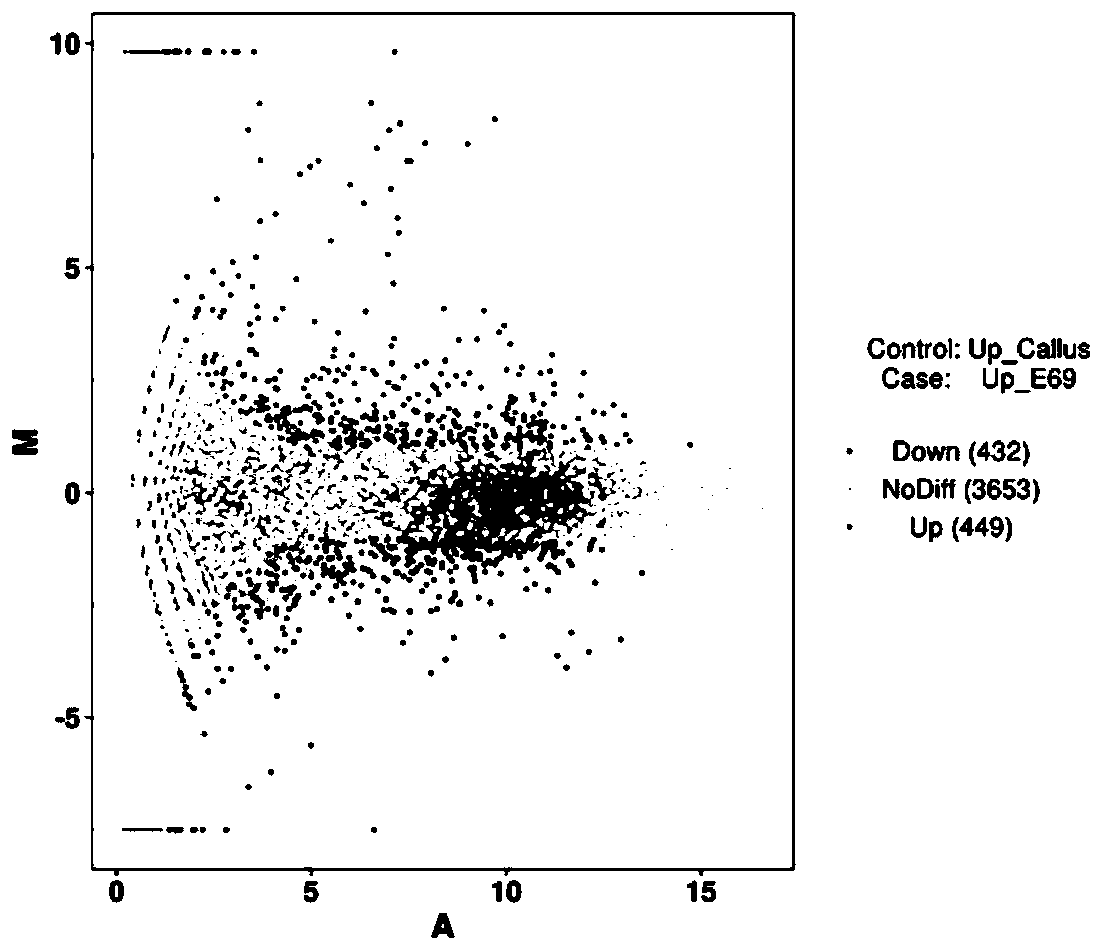
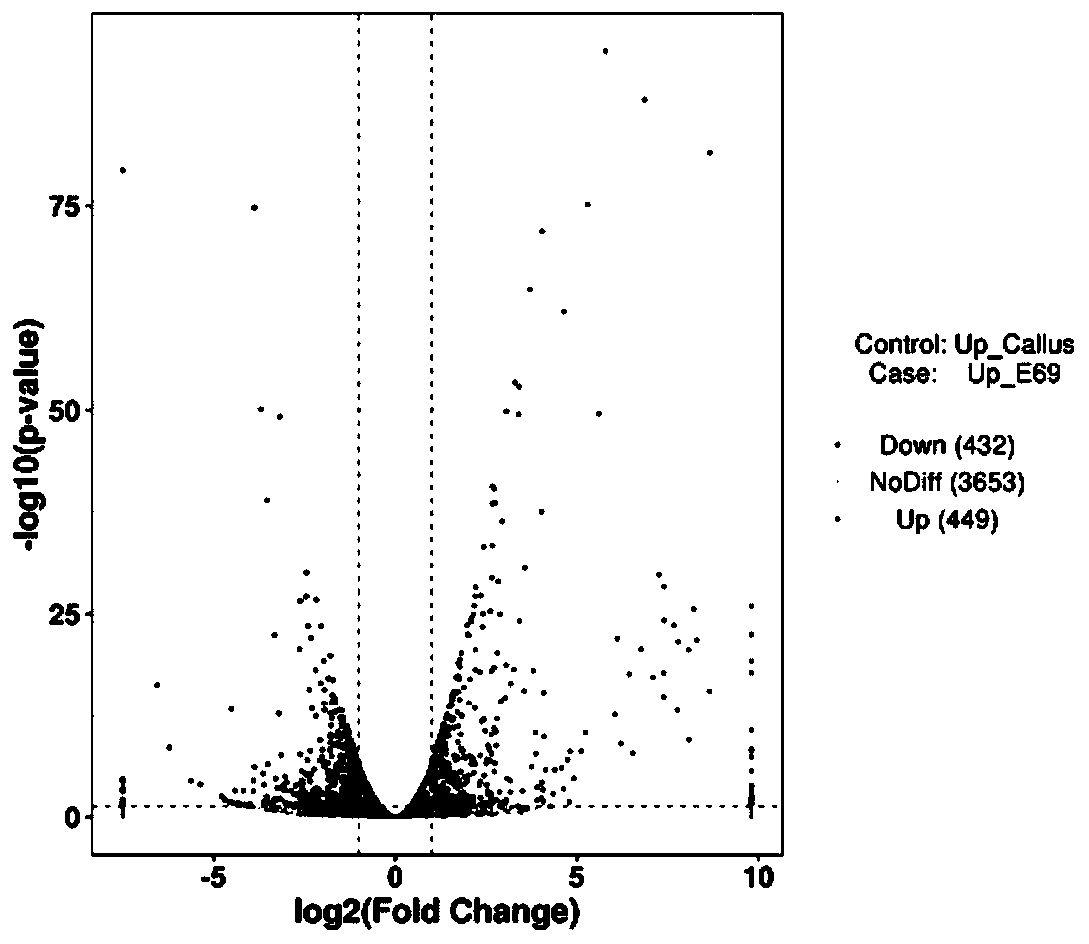
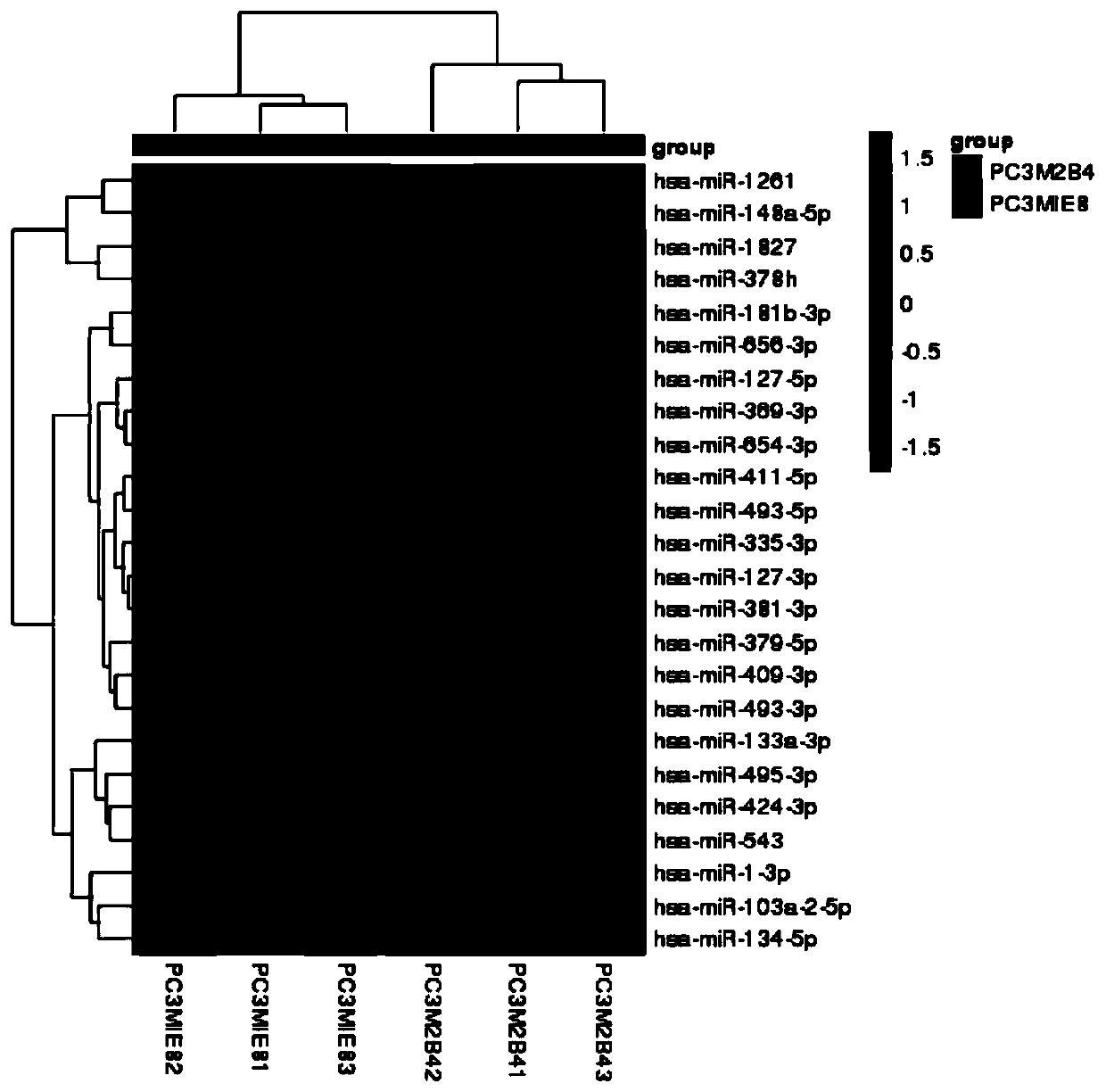
[0043] In step S1), the user's small RNA sequencing data and related database information are accepted, and then all data are analyzed to obtain annotation information of all small RNAs in each sample, and sequence characteristics analysis and expression of miRNAs are performed Quantitative analysis, as well as differential expression analysis between samples, function and pathway enrichment analysis.
[0044] The first is to filter and count the disembarkation data. In the embodiment of the present invention, the off-machine data is filtered to remove adapters and low-quality sequences, so as to obtain high-quality sequencing results. As an example, the perl language script is used to remove the linker sequence, and the origin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com