Method for analyzing iTRAQ (isobaric Tags for Relative and Absolute Quantitation) data
A data and database technology, applied in the field of quantitative data analysis of proteomics, can solve problems such as sensitivity and accuracy that are difficult to achieve ideal results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
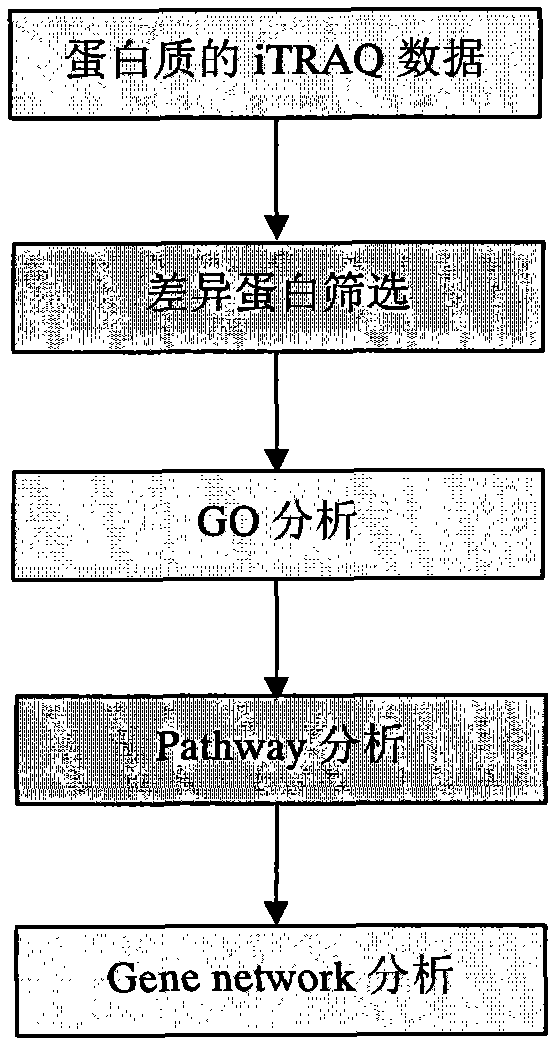
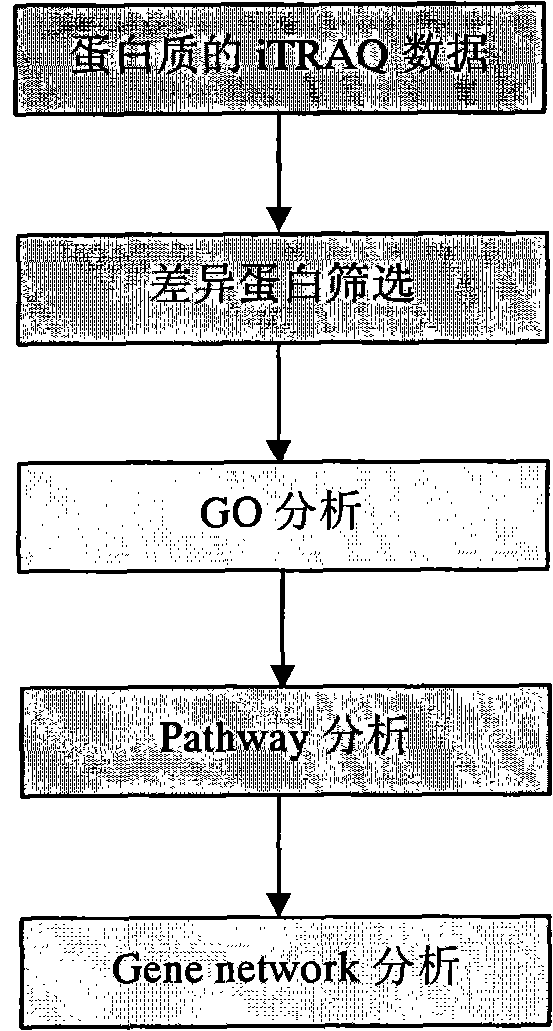
[0014] According to the present invention, a method for analyzing iTRAQ data is applied to the analysis of protein iTRAQ analysis data. In the present invention, the iTRAQ analysis data of mouse cells will be used as an example to illustrate the specific implementation steps of the method:
[0015] Step 1. Differential protein screening. The raw data of the screening came from the analysis results of the proteinpilot software after iTRAQ detection of mouse cells. The data were screened between groups according to the experimental group to obtain differentially expressed proteins. The follow-up analysis took the differential screening results of group C and group B in the experimental data as an example.
[0016] Step 2: GO (Gene ontology) analysis. The differentially expressed proteins obtained in the previous step can be used as input data for GO analysis, and the differentially expressed proteins are transferred to the Gene ontology database ( www.geneontology.org) to map ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com