Method for massively and efficiently developing molecular markers on basis of Indel and SSR (simple sequence repeat) site techniques
A molecular labeling and large-scale technology, applied in the fields of molecular biology and bioinformatics, can solve the problems of low effectiveness of SSR labeling and low efficiency of SSR molecular labeling, and achieve the effects of short time, low cost and low data volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The present invention will be described in detail below in conjunction with specific implementation methods.
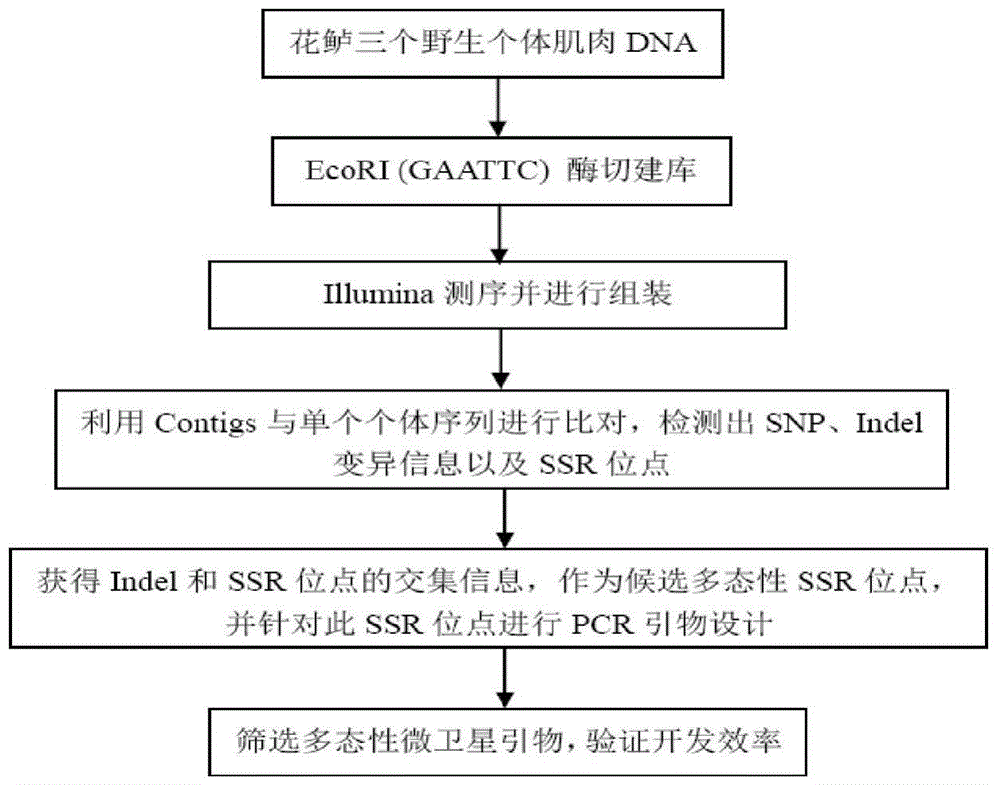
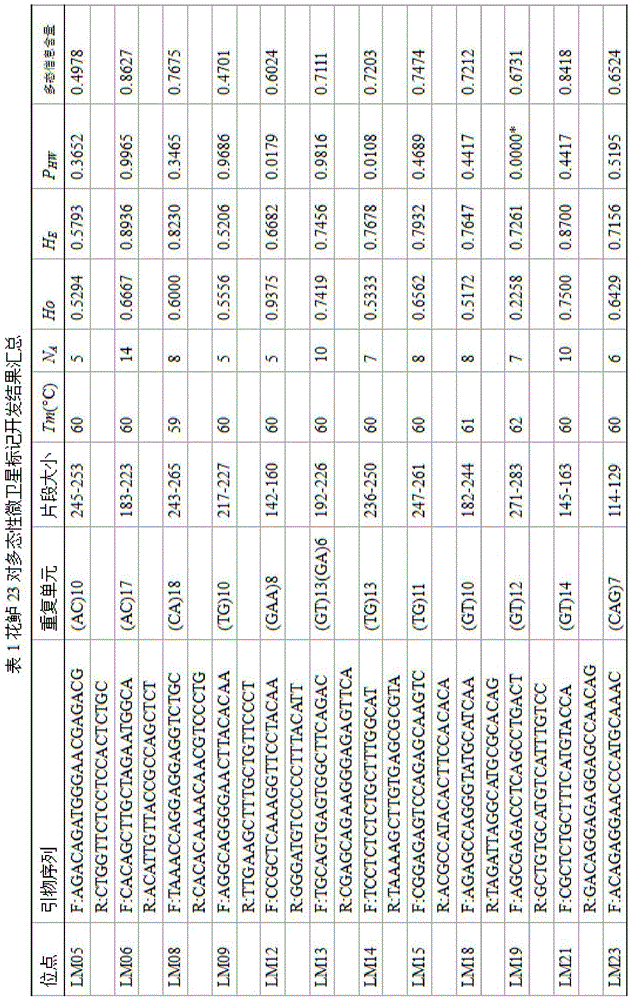
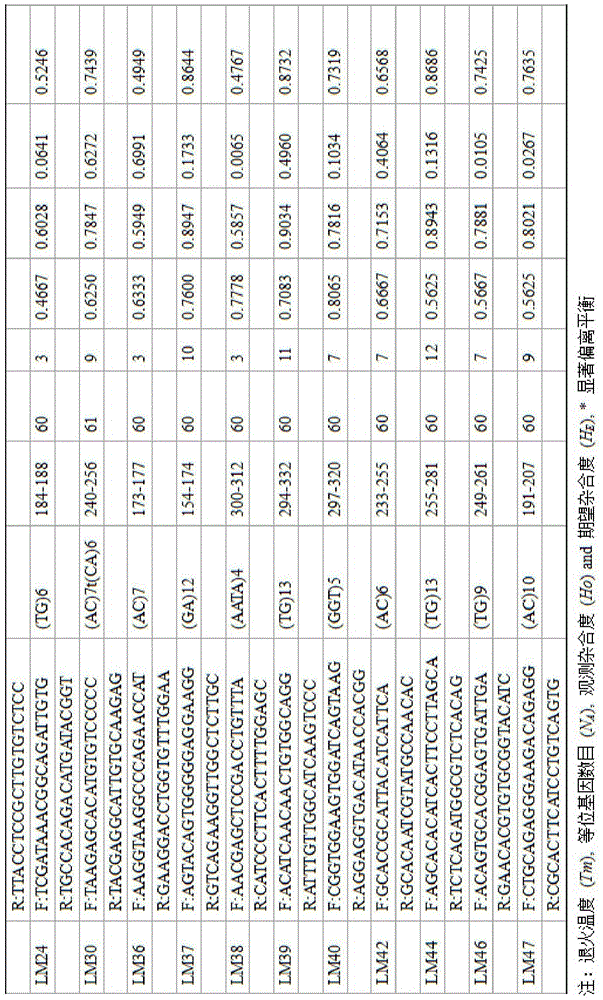
[0026] Such as figure 1 As shown, taking the perch as an example, a large-scale and efficient method for developing molecular markers for perch based on Indel and SSR locus technology includes the following steps:
[0027] (1) Extraction of genomic DNA from sea bass muscle samples
[0028] Select 3 wild perch muscle tissue samples, use the Magen animal tissue DNA extraction kit, process 20-50mg tissue samples into as small fragments as possible, and transfer them to a 1.5mL centrifuge tube. Add 550 μL BufferMTL and 20 μL Proteinase K, and vortex to mix. Incubate with shaking at 55°C for 3h or overnight to digest the samples. Add 5 μL RNaseSolution to the digestion solution, mix well by inverting. Stand at room temperature for 30-60min to digest RNA. Centrifuge at 13000xg for 3 min. Transfer the supernatant to a new 2.0 mL centrifuge tube. Add 500 μL Buff...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com