Method for identifying barley varieties by SSR (simple sequence repeat) primers and applications of method
A variety of barley technology, applied in the field of agricultural barley breeding and application, to achieve the effects of convenient operation, improved test efficiency, and less environmental impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Through analysis, 14 pairs of basic core SSR primers and 14 pairs of extended core SSR primers were identified among 153 pairs of SSR primers with polymorphism in barley varieties that have been published sequences:
[0028] The 14 pairs of basic core SSRs are Bmag0211, Bmag0378, Bmag0711, Bmac0209, Bmag0877, EBmac0701, EBmac0635, Bmag0353, Scssr10148, Scssr03907, Bmag0009, Bmag0867, HVCMA and EBmac0603.
[0029] The 14 pairs of extended core SSRs are Scssr02748, HVM54, Scssr07759, HvXan, HVM27, HVM60, HVM40, Bmac0181, Scssr07106, Bmag0387, Bmac0018, Scssr09398, Bmac0064 and Scssr15864.
[0030] For 14 pairs of basic core SSRs and 14 pairs of extended core SSRs, obtain the upstream primers and downstream primers corresponding to the SSRs through http: / / www.graingenes.org website (see Table 1):
[0031] Table 1 The primer sequence list is as follows:
[0032]
[0033] The primer sequences were synthesized by Shanghai Yingjun Biotechnology Co., Ltd.
Embodiment 2
[0035] The samples included two tested barley varieties, one of which was an unknown barley variety numbered B1 (hereinafter referred to as B1), and the other was a reference variety Supi No. 5. The two tested barley varieties were both produced in Jiangsu Province.
[0036] Reagent:
[0037] SuperTaq? DNA Polymerase (5U / μL), dNTP (2.5mmol / L), 10×PCR Buffer (containing Mg 2+ ); I set of SSR basic core primers synthesized by Shanghai Yingjun Biotechnology Co., Ltd.
[0038] Sampling and DNA extraction:
[0039] Random sampling of incoming samples. Take 5 individual plants for each sample, take 0.5 g of seedling leaves and mix them into one sample, cut them into pieces, put them into a 2 mL centrifuge tube, add liquid nitrogen to the centrifuge tube to freeze, put them into a grinder, and grind them into powder immediately Add about 800 μL of preheated DNA extraction solution (15 g SDS, 0.5 M EDTA (pH8.0), 1 M Tris-HCl (pH8.0), 5 M NaCl), shake vigorously to mix, and pla...
Embodiment 3
[0048] The steps and methods of this embodiment are the same as in Example 2, and the only difference between it and Example 2 is that one part of the sample is the unknown barley variety numbered B2 (hereinafter referred to as B2), and the other is the reference variety Su beer No. 3 (all places of origin are Jiangsu Province).
[0049] In the PCR reaction, 14 pairs of basic core SSR primers and 14 pairs of extended core SSR primers described in Table 1 were used to amplify the template DNA of the unknown barley variety B2 and the reference variety Supi No. 3.
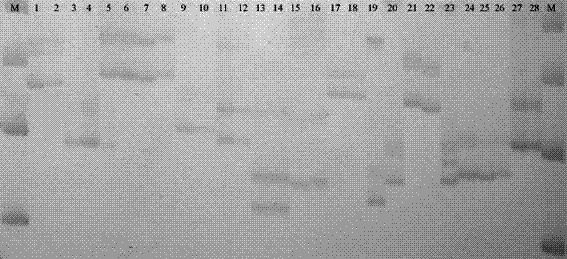
[0050] Denaturing polyacrylamide gel electrophoresis and silver staining electrophoresis figure 2 ,exist figure 2 Among them, the odd-numbered lanes are barley variety B2, and the even-numbered lanes are barley variety Supi No. 3. At the same time, lanes 1 and 2 are Bmag0211 primers, lanes 3 and 4 are Bmag0378, lanes 5 and 6 are Bmag0711, lanes 7 and 8 are Bmac0209 , lanes 9 and 10 are Bmag0877, lanes 11 and 12 ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com