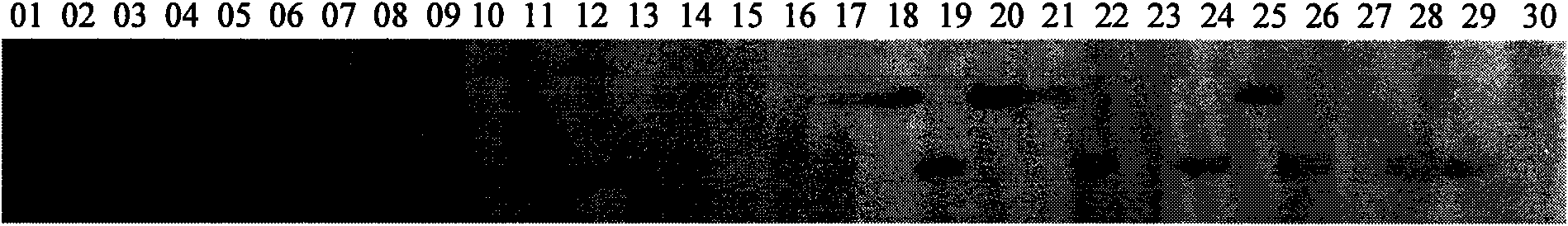Method for obtaining EST-SSR mark
A technology of labeling and simple sequence repetition, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as easy to miss SSR markers, miss SSR short sequences, polymorphism increase, etc. , achieve the effect of shortening R&D time, improving development efficiency and reducing development cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
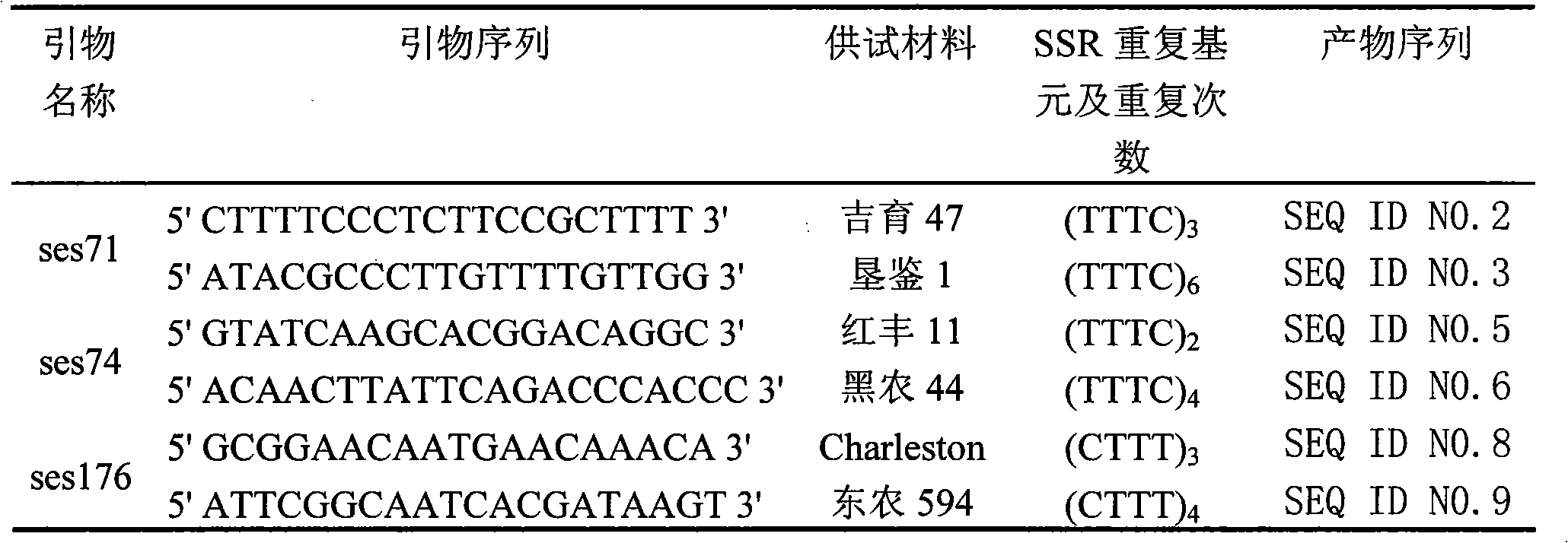
[0028] Example 1. Obtaining the EST-SSR marker of soybean
[0029] 1. Design of primers
[0030] 1. Obtain the EST sequence in the soybean genome
[0031] Soybean EST sequences were downloaded from the NCBI bioinformatics database, and a total of 458,220 soybean EST sequences were obtained.
[0032] 2. Search for EST sequences containing SSRs (ie simple sequence repeats)
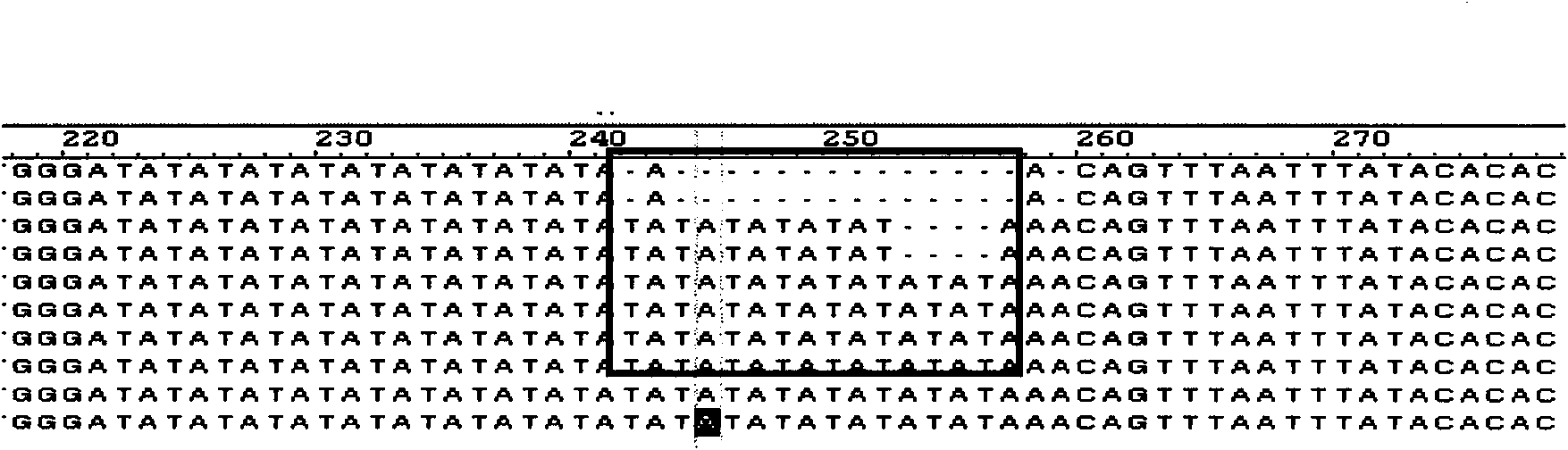
[0033]Use SSRIT (Simple Sequence Repeat Identification Tool) software to perform online search on all EST sequences obtained in step 1, and obtain EST sequences containing simple repeat sequences (called SSR-EST sequences); the search criteria are: dinucleotides, trinucleotides The number of repetitions of the nucleotide, tetranucleotide, pentanucleotide and hexanucleotide repeating units is greater than or equal to 6, 5, 4, 4, and 4, respectively.
[0034] 3. Classify SSR-EST sequences
[0035] According to the different types of simple sequence repeating units, all SSR-EST sequences obtained in step 2 ...
Embodiment 2
[0159] Example 2: Comparison between the EST-SSR marker development method of the present invention and conventional methods
[0160] Existing method: From all the EST-SSR sequences obtained in Example 1, 191 sequences were randomly selected for primer development and design, and a pair of primers was obtained for each sequence, and a total of 191 pairs of primers were obtained. Then, the polymorphism of each pair of primers was detected by the method described in Experiment 2 in Example 1, and the ratio of the polymorphic primers was counted. The results are shown in Table 7. Ratio of polymorphic primers=number of polymorphic primers / number of all primers.
[0161] Table 7. EST-SSR polymorphism statistics of existing methods
[0162]
[0163] Comparing the research results in Table 5 and Table 7, it can be seen that the common feature of the two methods is that the ratio of polymorphic primers generated by 3'EST is higher than that of 5'EST and EST from other sourc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com