Seed number per pod character major gene site of rape and application thereof
A technology of main effect genes and loci, which is applied in the field of molecular biology and genetic breeding, can solve the problems of small repeatability of QTL effect value, difficulty in the application of rapeseed breeding, etc., and achieve convenient and fast detection methods, accurate and fast screening, The effect of saving production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
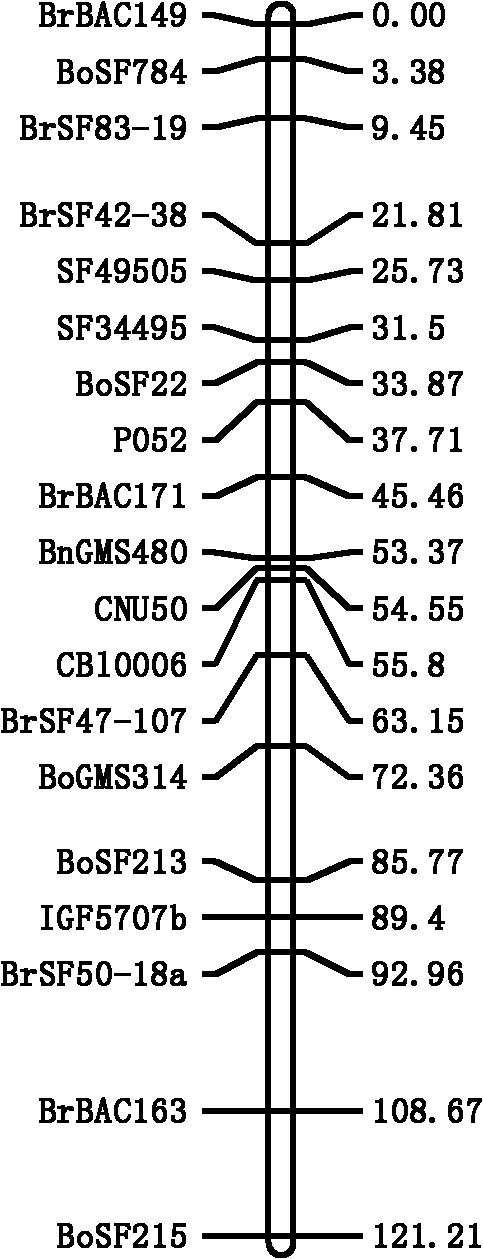
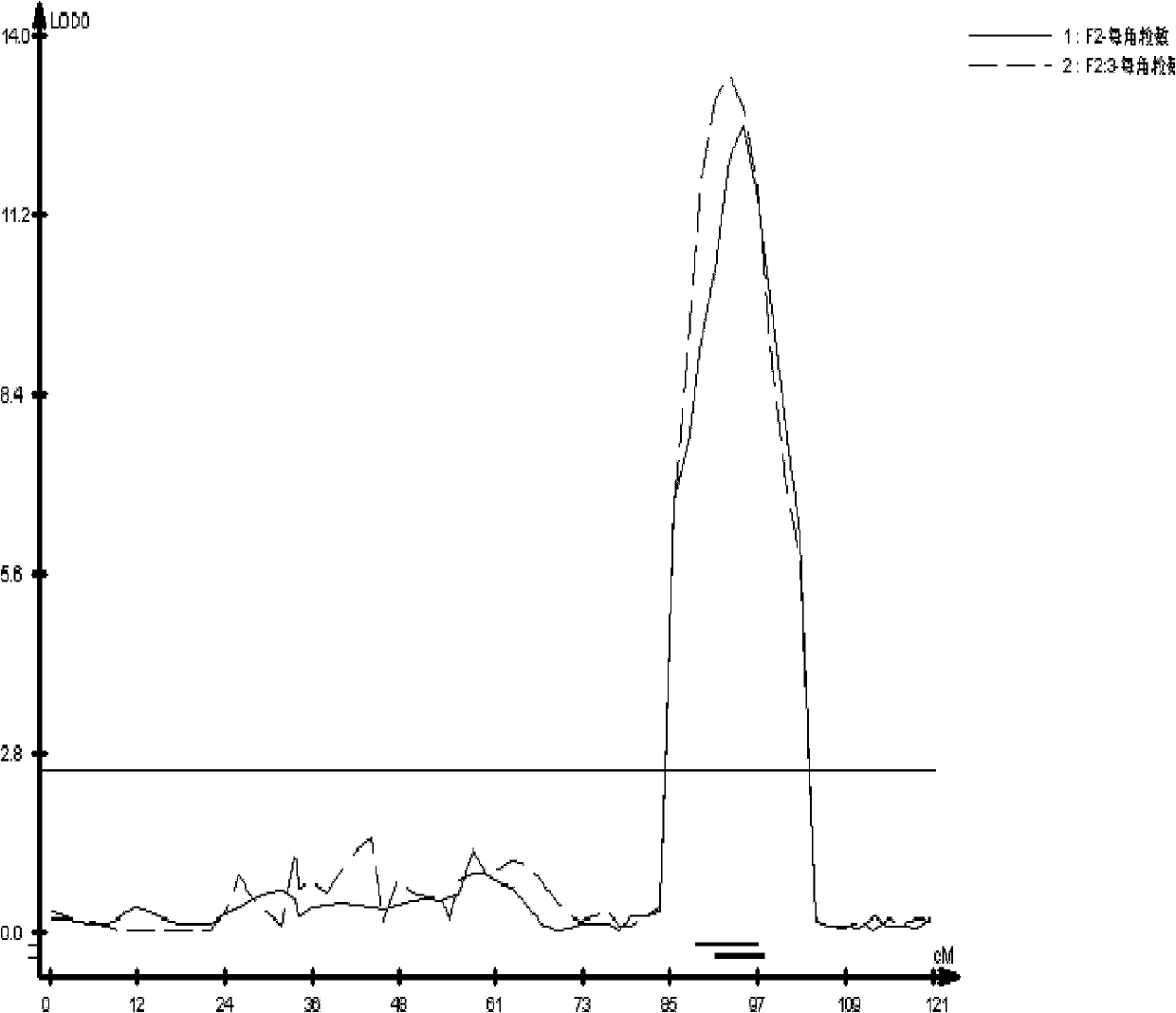
[0033] Construction and trait determination of rapeseed segregated populations per corner:
[0034] The isolated populations used in this example are the F2 and F2:3 populations derived from Shuang11 (≈21 grains) and 73290 (≈11 grains) of rapeseed with many and few grains. The phenotypes of the number of seeds per corner of the two parents and the two populations were identified after harvesting at the maturity stage. The number of seeds per corner species test data shows that the two parents have weak super-parental segregation, indicating that the multigrain gene is mainly distributed in the genome of Zhongshuang 11; the number of seeds per corner of the two populations is normally distributed, proving the trait of number of seeds per corner Quantitative genetic characteristics of ( figure 1 ).
Embodiment 2
[0036] Extraction of total DNA from leaves:
[0037] The total DNA of leaves was extracted by the CTAB method, and the specific steps were as follows:
[0038] (1) Take 0.1 g of fresh leaves and grind them, add 700 microliters of extract to grind them, then transfer them to a 1.5 ml centrifuge tube and place them in a constant temperature water bath at 65°C for 60 minutes, during which they mix 2-3 times;
[0039] (2) Add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1, V / V / V), invert gently to mix well, centrifuge at 12000rpm for 10 minutes, and gently suck the supernatant into another 1.5 ml centrifuge tube; add an equal volume of chloroform:isoamyl alcohol (24:1, V / V) and re-extract once;
[0040] (3) Add 1 ml of -20°C pre-cooled absolute ethanol, and freeze at -20°C for no more than 30 minutes to precipitate DNA;
[0041] Centrifuge at 12000rpm for 10 minutes to allow DNA to precipitate, discard the ethanol solution in the centrifuge tube; wash 2-3 times ...
Embodiment 3
[0044] Primer development and synthesis:
[0045] The SSR primers utilized by the applicant include two types: one is the primer sequences published in published articles and Brassica databases (http: / / www.brassica.info / resource / markers / ssr-exchange.php); the other The series were developed by the applicant based on the sequences of Chinese cabbage and cabbage scaffolds, named BrSF and BoSF series respectively. The specific development method is to use SSRHunter software to search for SSRs in each scaffold, and then use Primer3.0 software to design SSR primers. The self-developed SNP primers are obtained by comparing the resequencing sequence of 73290 with the reference genome sequence of Zhongshuang11. First, the resequencing sequence of 73290 is positioned on the whole genome reference sequence of Zhongshuang11 using BWA software, and then Use samtools software to find SNP. The SNP detection adopts the SNAP (single nucleotide amplified polymorphism) method, that is, a misma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com