Rape pod shatter relevant gene, molecular marker and application
A gene and rapeseed technology, which is applied to the related genes and molecular markers of rapeseed angle and in the fields of crop genetic breeding and physiology, can solve the problems of unutilized key genes and functional molecular markers, and achieves speeding up the rapeseed breeding process and screening. The effect of progress and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
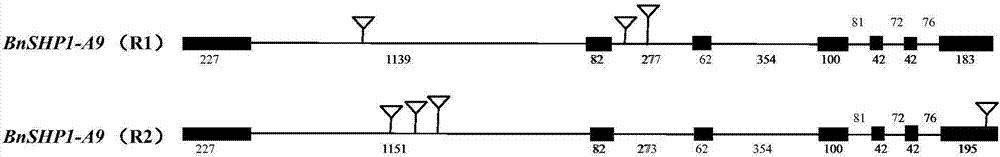
[0039] Example 1: Acquisition of rapeseed horn-related gene BnSHP1-A9:
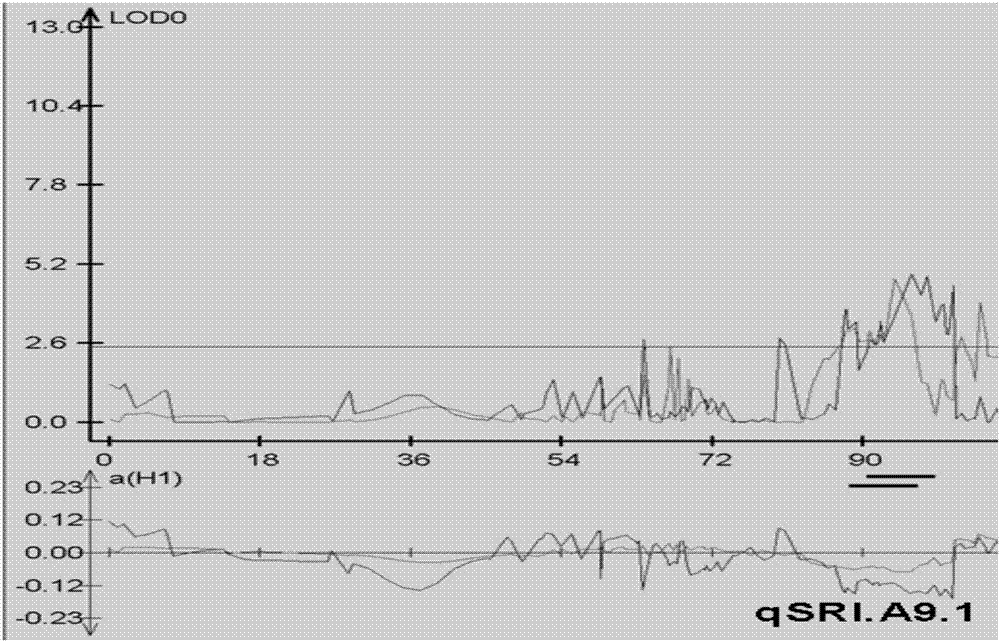
[0040] 1. Location and cloning of qSRI.A9.1
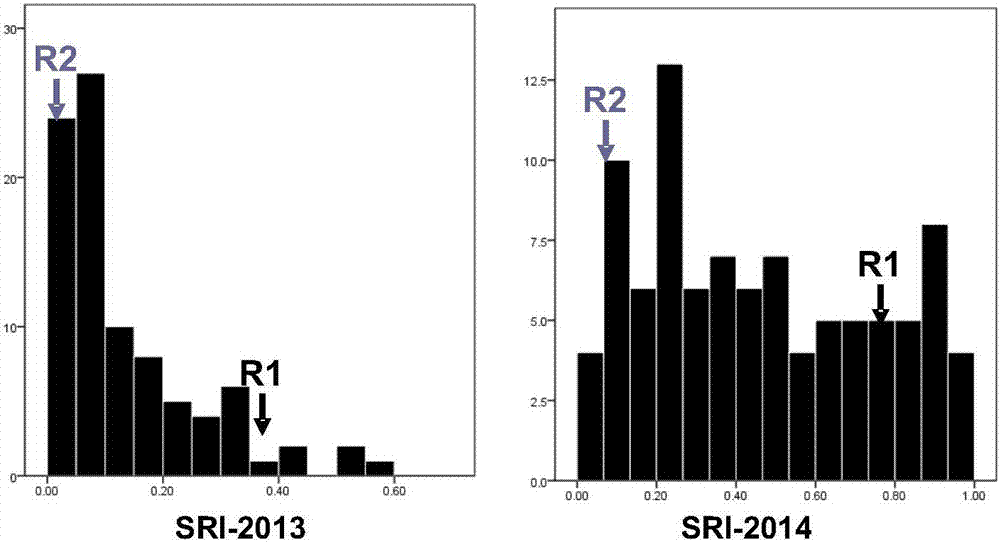
[0041] (1) Field experiment and phenotypic determination of anti-cracking angle index:
[0042] Field experiments were conducted at the Yangluo Experimental Station (Wuhan) of the Oil Plant Research Institute of the Chinese Academy of Agricultural Sciences from 2013 to the autumn of 2014. The crack angle resistance index (SRI) of the strains or individual plants was investigated at the mature stage. The identification method refers to patent 201310226680.4. Specifically, the rape siliques were dried at 80°C for 30 minutes, sealed and stored at room temperature overnight, then placed in a cylindrical container, and 8 steel balls were placed in the cylindrical container; the cylindrical container was shaken at a speed of 280rpm using a model HQ45Z shaker. Observe once every two minutes, observe 5 times in total, repeat 3 times for each material, use the formula...
Embodiment 2
[0056] Example 2: Differential expression analysis of BnSHP1-A9 was carried out by sampling the corresponding parts of the R1 and R2 siliques at different developmental stages.
[0057] From different flower buds to flowering, until the formation of siliques at different stages of development, taking the initial stage of rapeseed flowering as the standard, sampling every 2 days after the formation of siliques 10 days after pollination, according to the development period of siliques, each material in each period is parallel Five plants with uniform development and size of siliques were selected as five biological replicates, and the cut siliques contained the same proportion of detached tissues as much as possible. Use RNAprep pure Plant Kit (TIANGEN) to extract total RNA, and then use Hiscript @ II 1 st The strandcDNA synthesis kit reverse-transcribes RNA into first-strand cDNA, using 2×Es Taq MasterMix (CWBIO), BIO-RAD S1000 TM Thermal Cycler instrument for RT-PCR reacti...
Embodiment 3
[0059] Example 3: Construction of Rapeseed BnSHP1-A9-R1 Suppression Expression Vector and Transformation in Rapeseed
[0060] According to the BnSHP1-A9 coding region sequence of R1 material, primers were designed to amplify part of the coding region fragment (143bp). The primer sequences were: TTCTTTTCGGTGGTTTTATTC and TGACGATTTGTTGTGTTCTCT. It was reverse ligated to the topo vector and recombined into pEarleyGate100. Transformation of the vector into LBA4404 is ready for transformation of canola.
[0061] Transformation of Rapeseed with Agrobacterium LBA4404
[0062] A. Preparation of sterile seedlings: Seeds were soaked in 70% ethanol for 1 min, mercuric chloride (HgCl 2 ) for 13-15 min, ddH 2 O washed 5 times, and spread in MS medium (PH 5.8), agar concentration 0.8%. Rapeseed aseptic seedlings for use.
[0063] B. Rapeseed petiole transformation: inoculate Agrobacterium tumefaciens LBA4404 on solid medium LB, pick a single colony after two days and cultivate in 50ml YE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com