Cucumber variety identification method based on SNP markers
A variety, cucumber technology, applied in the field of agricultural vegetable breeding and application, can solve problems such as affecting the yield and income of growers, insufficient marking density, complicated operation, etc. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
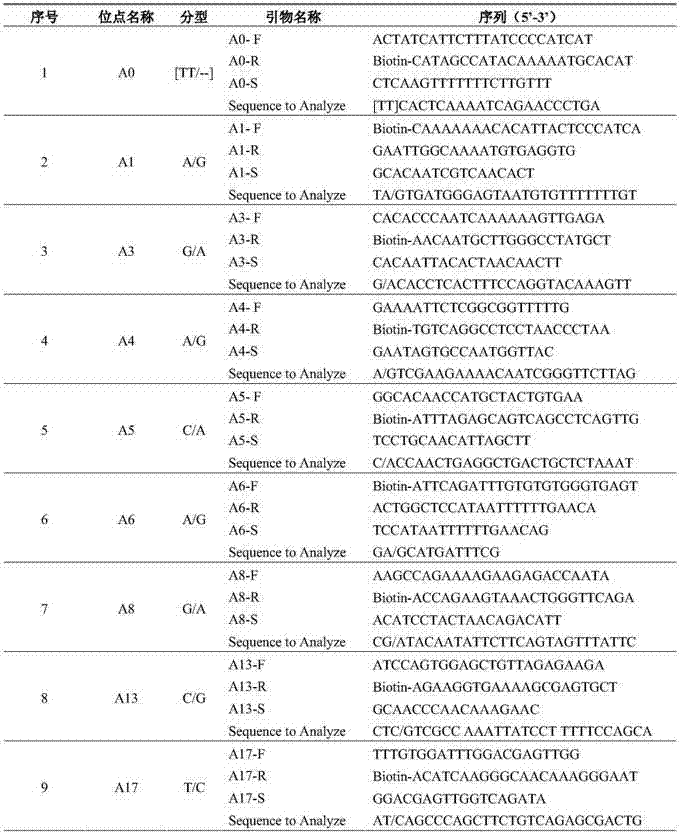
[0040] (1) Reagent: GoTaq produced by Promega Master Mix solution; specific amplification primers and sequencing primers synthesized by Shanghai Sangon; Sepharose Bead produced by Biotage.
[0041] (2) Amplification reaction system and amplification program: the total volume of the amplification reaction is 50 μL, and its various components are: 2×GoTaq 25 μL of Master Mix, 1 μL of 10 μmol / L primers, 2 μL of DNA templates (cucumber seed DNA) of the test variety and the control variety, make up to 50 μL with sterilized deionized water; reaction program: 95 °C pre-denaturation for 10 min, 94 °C denaturation for 30 seconds , annealed at 50°C for 30S, extended at 72°C for 45S, cycled 50 times, finally extended at 72°C for 7min, and stored at 4°C.
[0042] (3) Sequencing reaction system: Add 47 μL Binding buffer and 3 μL Sepharose beads to 50 μL PCR product, vortex and mix at 1300 rpm for 15 minutes, separate the single strands by Vacuum prep workstation, and release them into t...
Embodiment 2
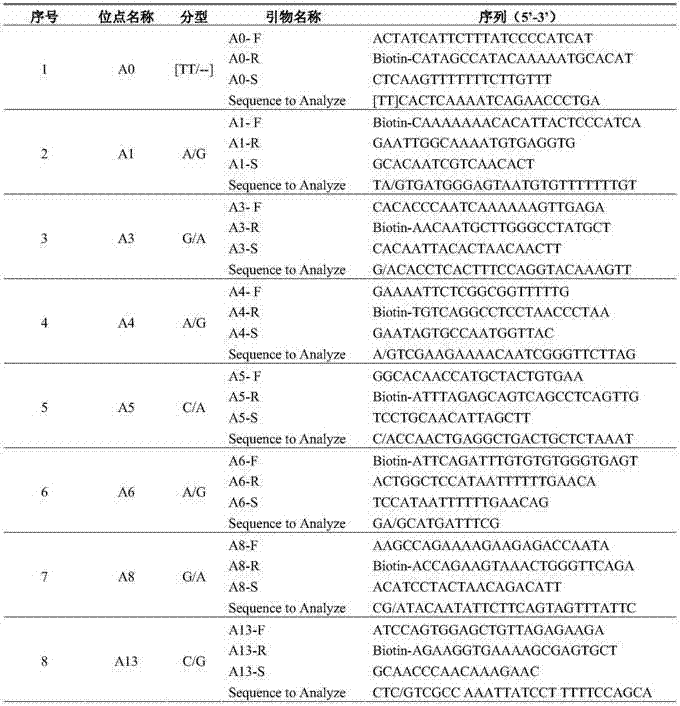
[0047] (1) Reagent: GoTaq produced by Promega Master Mix solution; specific amplification primers and sequencing primers synthesized by Shanghai Sangon; Sepharose Bead produced by Biotage.
[0048] (2) Amplification reaction system and amplification program: the total volume of the amplification reaction is 50 μL, and its various components are: 2×GoTaq 25 μL of Master Mix, 1 μL of each 10 μmol / L primer, 2 μL of template DNA (cucumber seed DNA) of the test variety and control variety, filled to 50 μL with sterilized deionized water; Anneal at 50°C for 30S, extend at 72°C for 45S, cycle 50 times, finally extend at 72°C for 7min, and store at 4°C.
[0049] (3) Sequencing reaction system: Add 47 μL Binding buffer and 3 μL Sepharose beads to 50 μL PCR product, vortex and mix at 1300 rpm for 15 minutes, separate the single strands by Vacuum prep workstation, and release them into the PSQ pre-added with 38.8 μL Annealing buffer and 1.2 μL sequencing primers In the 96 sequencing ...
Embodiment 3
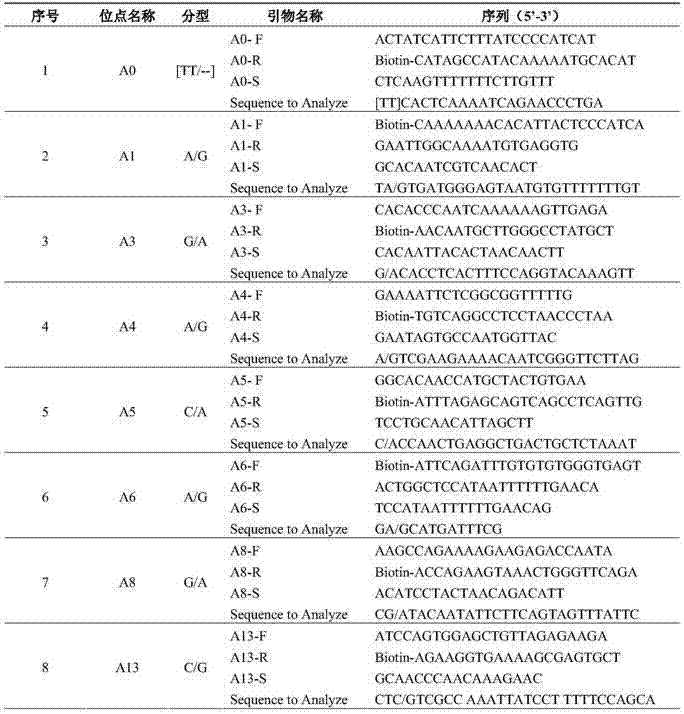
[0054] (1) Reagent: GoTaq produced by Promega Master Mix solution; specific amplification primers and sequencing primers synthesized by Shanghai Sangon; Sepharose Bead produced by Biotage.
[0055] (2) Amplification reaction system and amplification program: the total volume of the amplification reaction is 50 μL, and its various components are: 2×GoTaq 25 μL of Master Mix, 1 μL of each 10 μmol / L primer, 2 μL of template DNA (cucumber seed DNA) of the test variety and control variety, filled to 50 μL with sterilized deionized water; Anneal at 50°C for 30S, extend at 72°C for 45S, cycle 50 times, finally extend at 72°C for 7min, and store at 4°C.
[0056] (3) Sequencing reaction system: add 47 μL Binding buffer and 3 μL Sepharose beads to 50 μL PCR product, vortex and mix at 1300 rpm for 15 minutes, separate the single strands by Vacuum prep workstation, and release them into PSQ96 pre-added with 38.8 μL Annealing buffer and 1.2 μL sequencing primer In the sequencing reacti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com