Primers, method and kit for detecting secreted frizzled-related protein (SFRP)2 gene CpG island methylation
A technology of SFRP2-F and SFRP2-R, which is applied in the fields of life science and biology, can solve the problem of high probe ordering costs and achieve high accuracy and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Primers and kits for detection of methylation of SFRP2 gene CpG island
[0044] Primers for detecting methylation of SFRP2 gene CpG islands, including specific primers for amplifying SFRP2 gene fragments from sample nucleic acid and primers for pyrosequencing the obtained nucleic acid fragments, wherein,
[0045] Specific primers for amplifying the CpG island DNA fragment of the gene SFRP2 gene from the sample nucleic acid, the base sequence of which is:
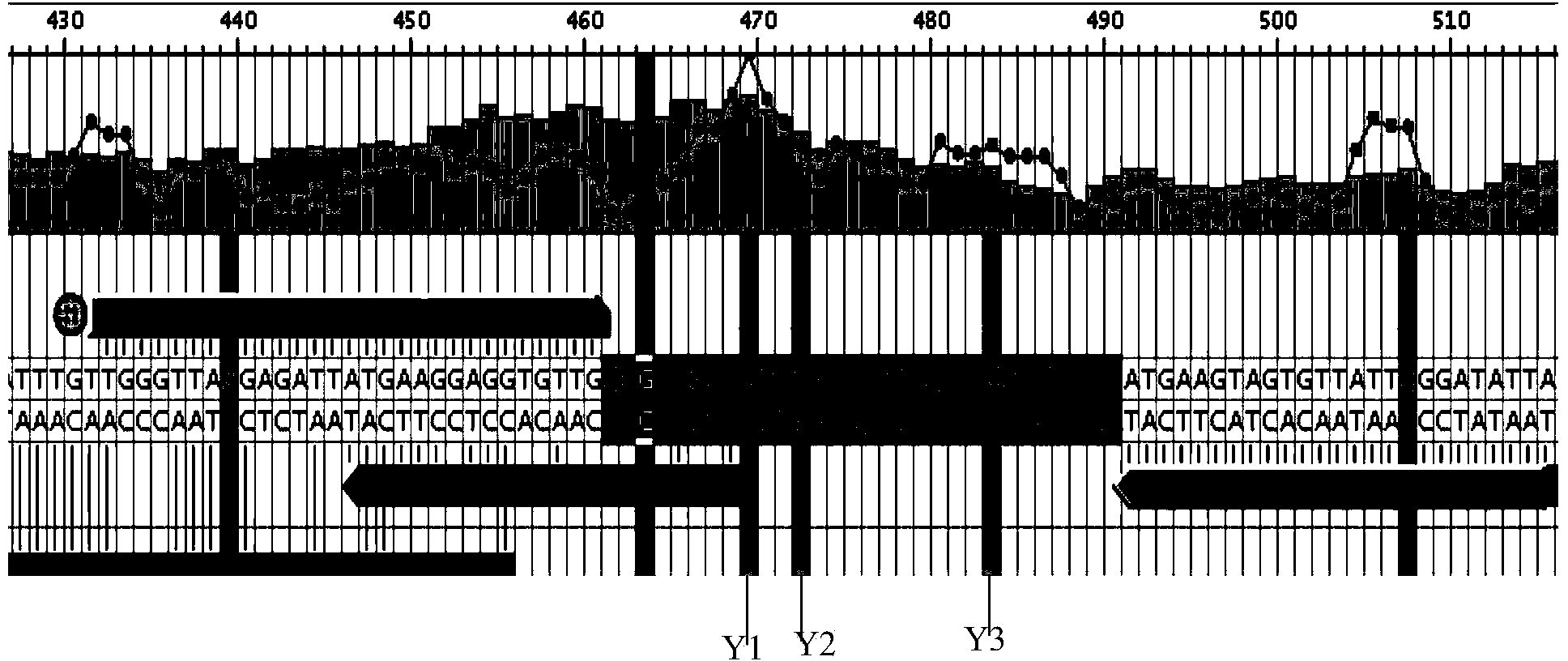
[0046] SFRP2-F:Biotin-TGGGTTAAGAGATTATGAAGGAGGTGTTG
[0047] SFRP2-R:CTCAACCCAACAAAAAATAAAAAAAACAA
[0048] Wherein SFRP2-F is the 5' end biotin-labeled primer
[0049] The obtained nucleic acid amplification fragment is subjected to pyrosequencing primers, and its base sequence is:
[0050] SFRP2-S: AATATCCCAATAACACTACTTCAT (reverse).
[0051] Using pyrosequencing technology, through specific amplification primers SFRP2-F and SFRP2-R and pyrosequencing primers, the CG methylation of CpG islands related...
Embodiment 2
[0062] Example 2: DNA Extraction
[0063] DNA extraction reagents: stool suspension, stool DNA extraction kit (TIANGEN company).
[0064]Operation steps: Weigh 180-220mg of stool sample into a 2ml centrifuge tube, place the tube on ice and add 0.5ml of stool suspension, then add 1.4ml buffer GSL to the sample, shake intermittently for 1 minute until the sample is well mixed , incubate at 70°C for 5 minutes, vortex for 15 seconds, and centrifuge at 13,000 rpm for 1 minute. Transfer 1.2ml of the supernatant to a new 2ml centrifuge tube, add an inhibitor adsorption sheet InhibitEX, oscillate until the adsorption sheet is completely opened and resuspended, incubate at room temperature for 1 minute, so that the adsorption sheet can fully function, then centrifuge at 13000rpm for 3 minutes, and the obtained Transfer the supernatant to a new 1.5ml centrifuge tube, centrifuge at 13000rpm for 3 minutes, transfer the obtained supernatant 200μl to a new 1.5ml centrifuge tube, add 15μl p...
Embodiment 3
[0065] Embodiment 3: DNA sodium bisulfite treatment
[0066] After DNA was treated with sodium sulfite, the unmethylated cytosine in the CpG dinucleotide pair was converted to thymine (C→T), while the methylated 5'methylcytosine was not affected by sodium sulfite treatment, Still 5'methylcytosine.
[0067] Operation steps: Dilute the genomic DNA extracted in Example 2 and 1.5ml EP tube to 50ul with double distilled water, add 5.5ul freshly prepared 3M NaOH solution, and bathe in water at 42°C for 30min; prepare the following solution during the water bath: 10mM hydroquinone solution (Hydryquinone) : Dissolve 55mg hydroquinone in 50ml water; 3.6mol / LNaHSO 3 : Take 18.8g NaHSO 3 Add 40ml of water, adjust the pH of the solution to 5.0 with 3M NaOH solution, and finally dilute to 50ml. Add 30ul10mM hydroquinone solution and 520ul3.6mol / LNaHSO 3 Put the EP tube into the solution after the water bath, wrap the EP tube with aluminum foil to avoid light, gently invert the solution...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com