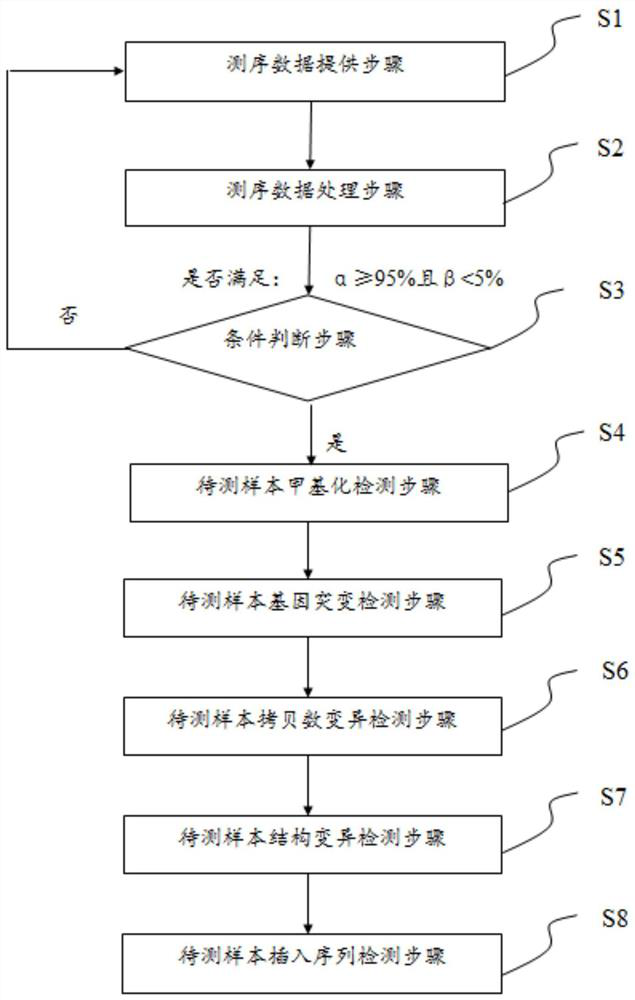
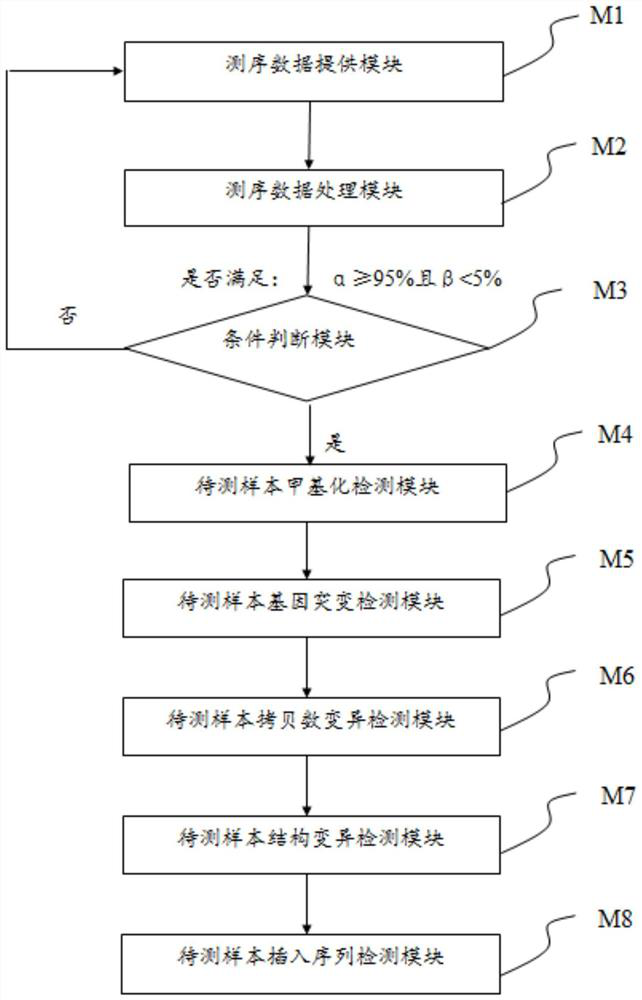
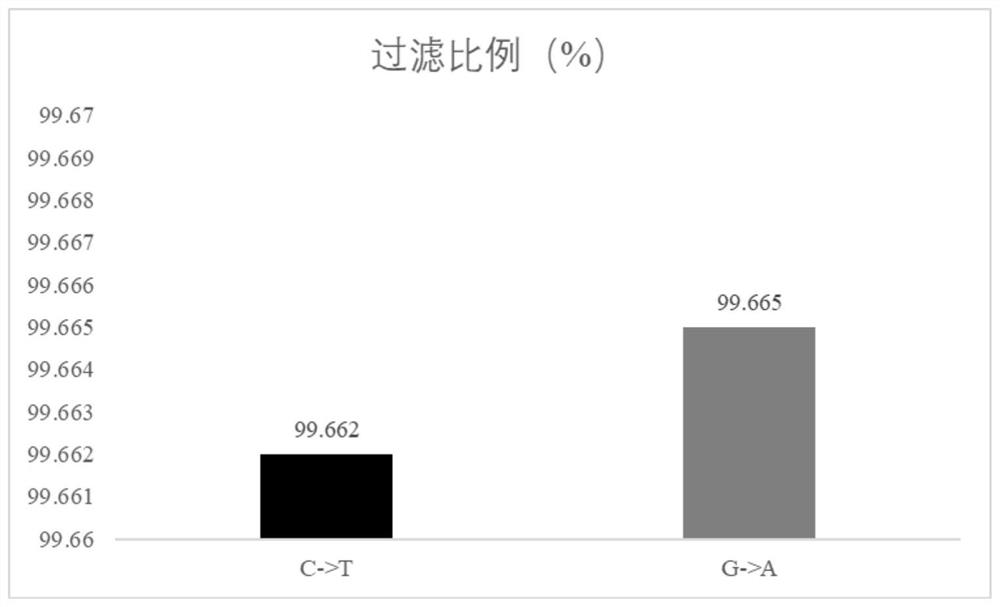
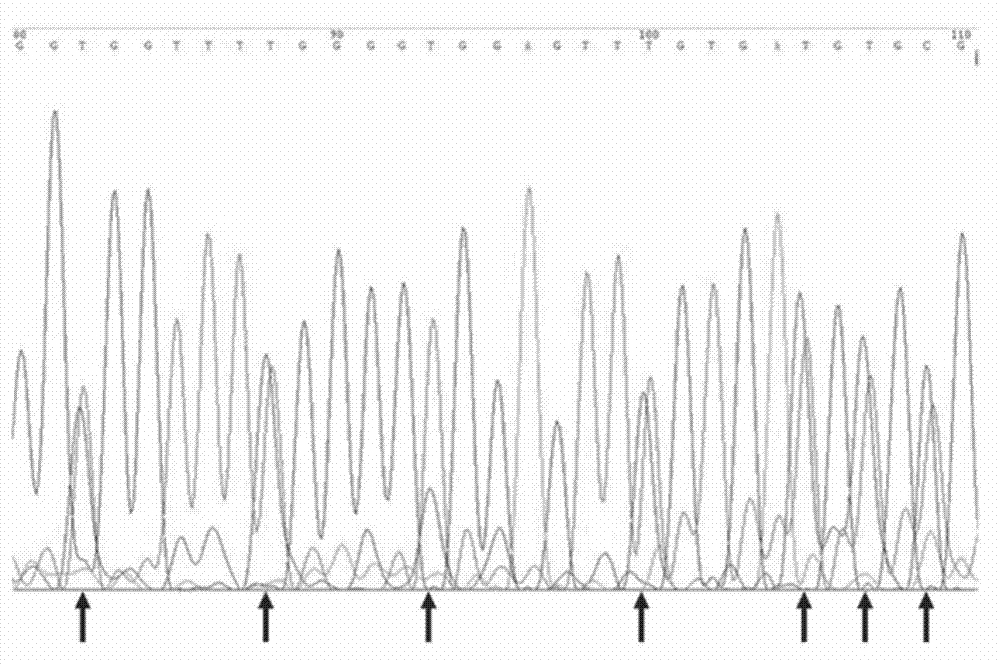
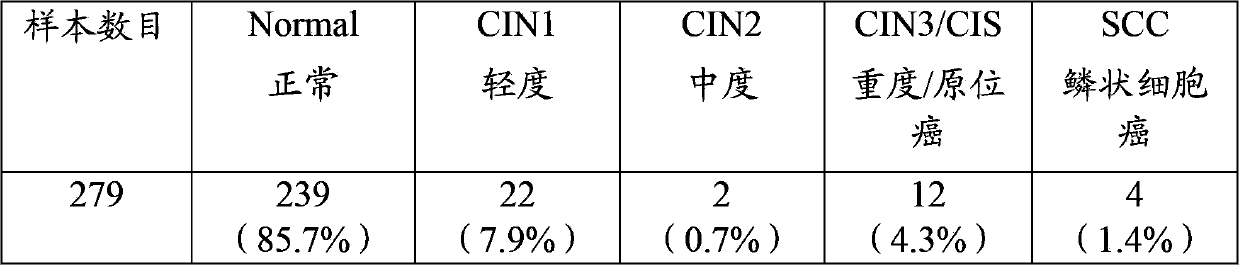
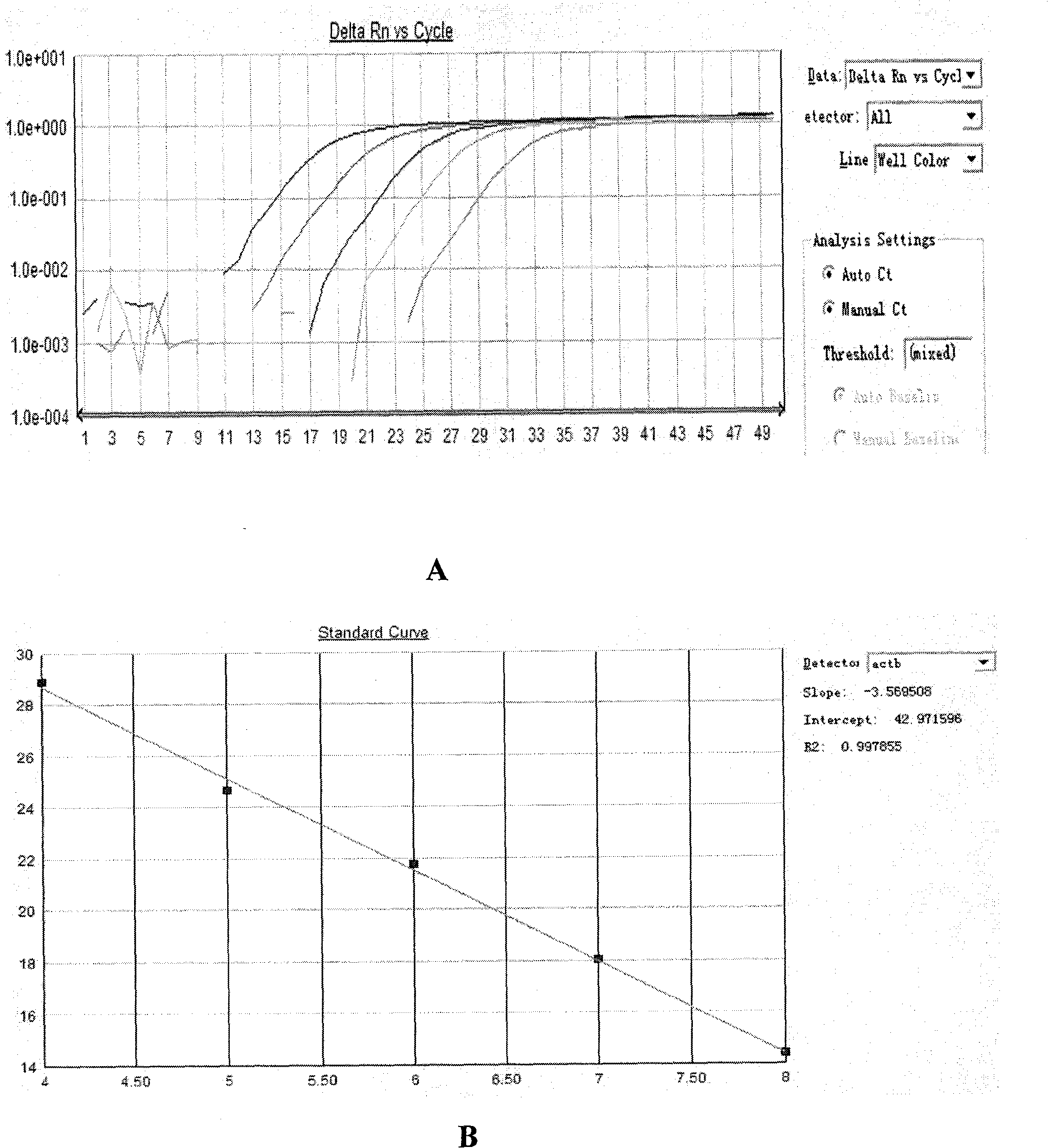
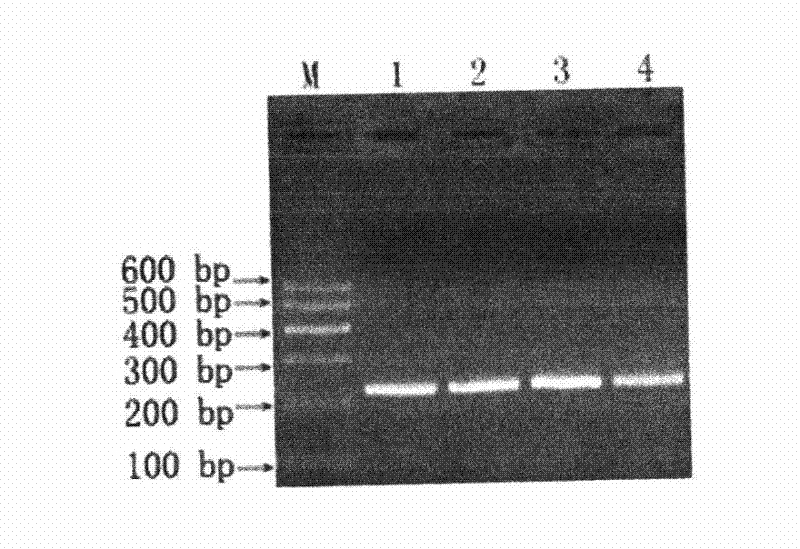
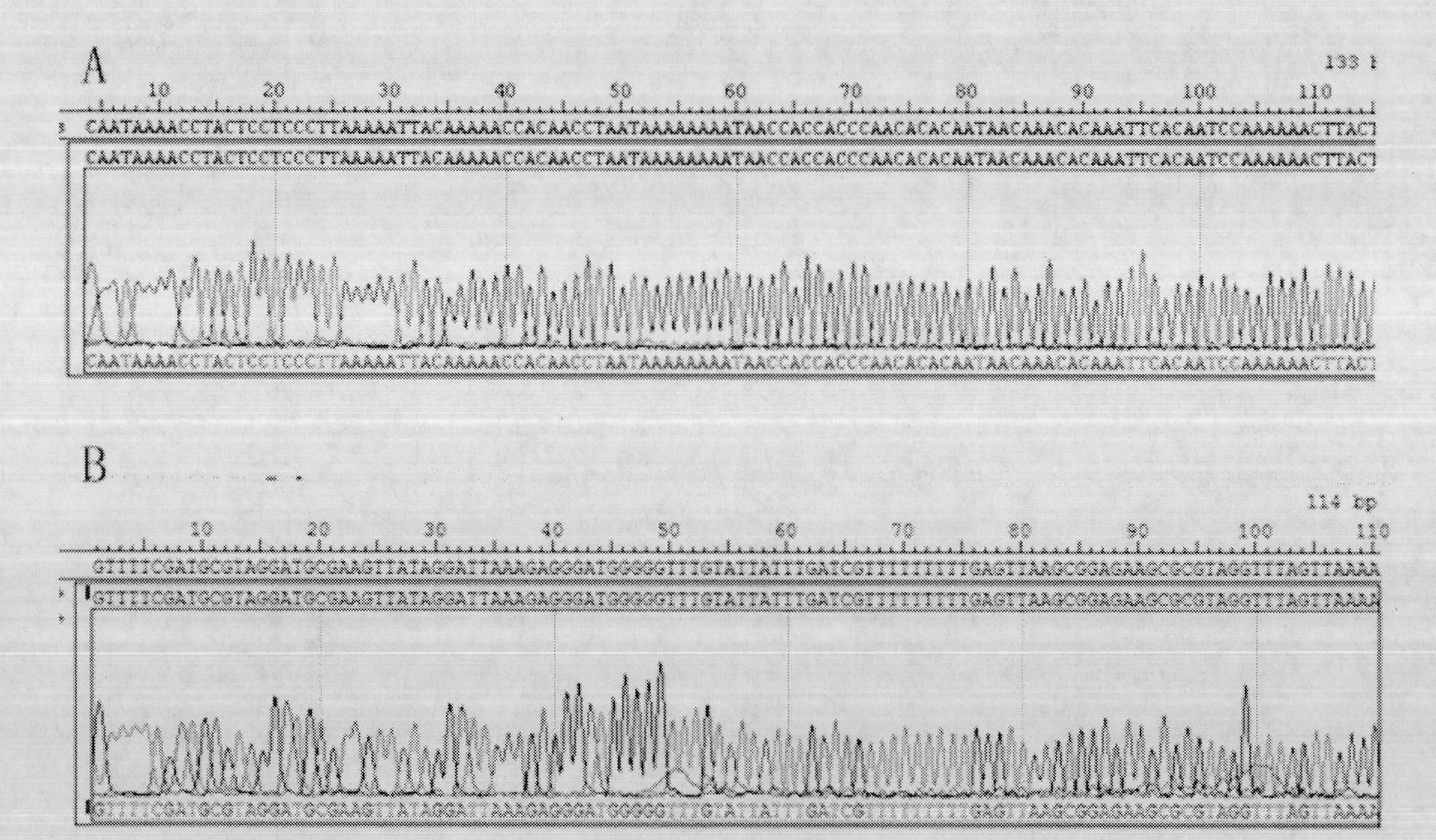
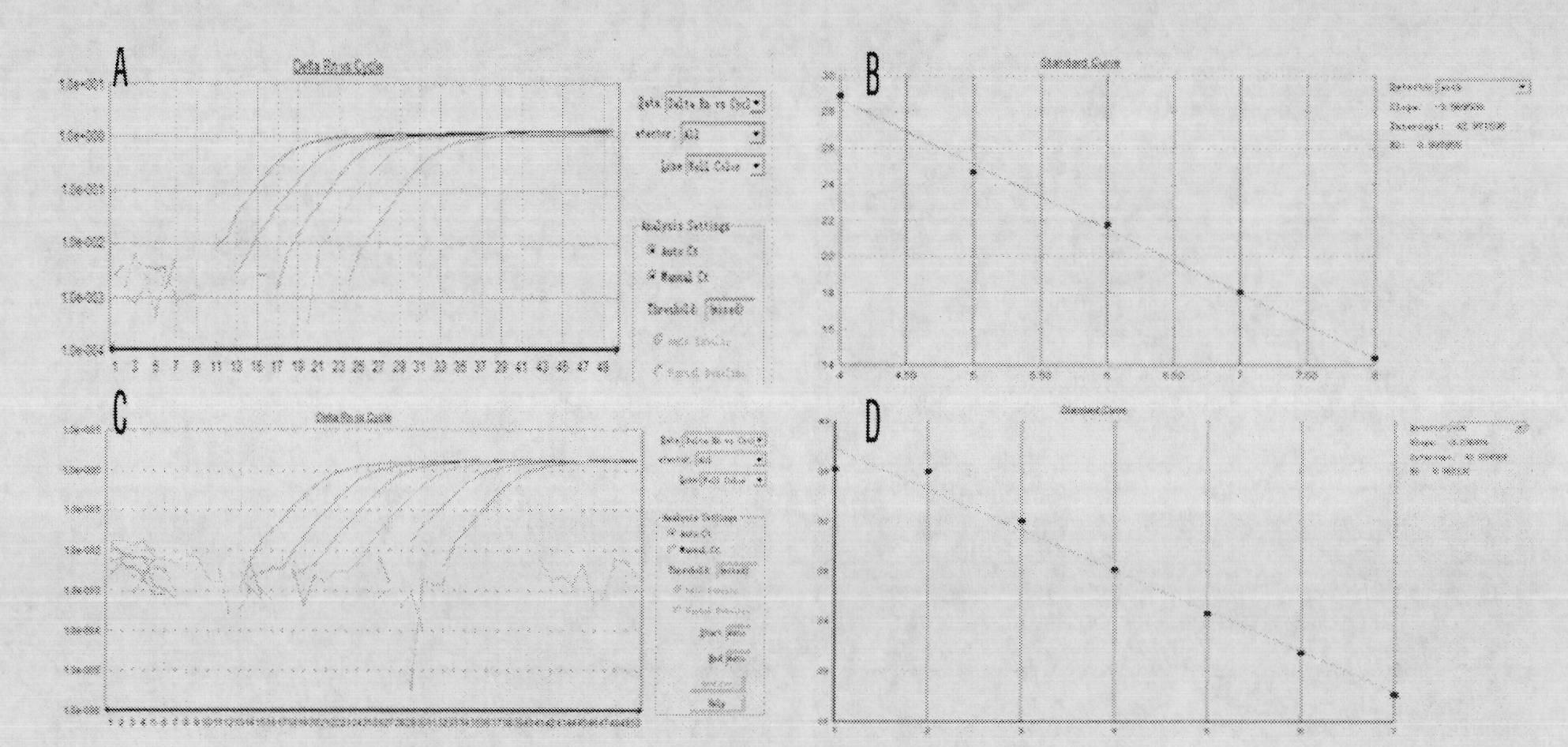
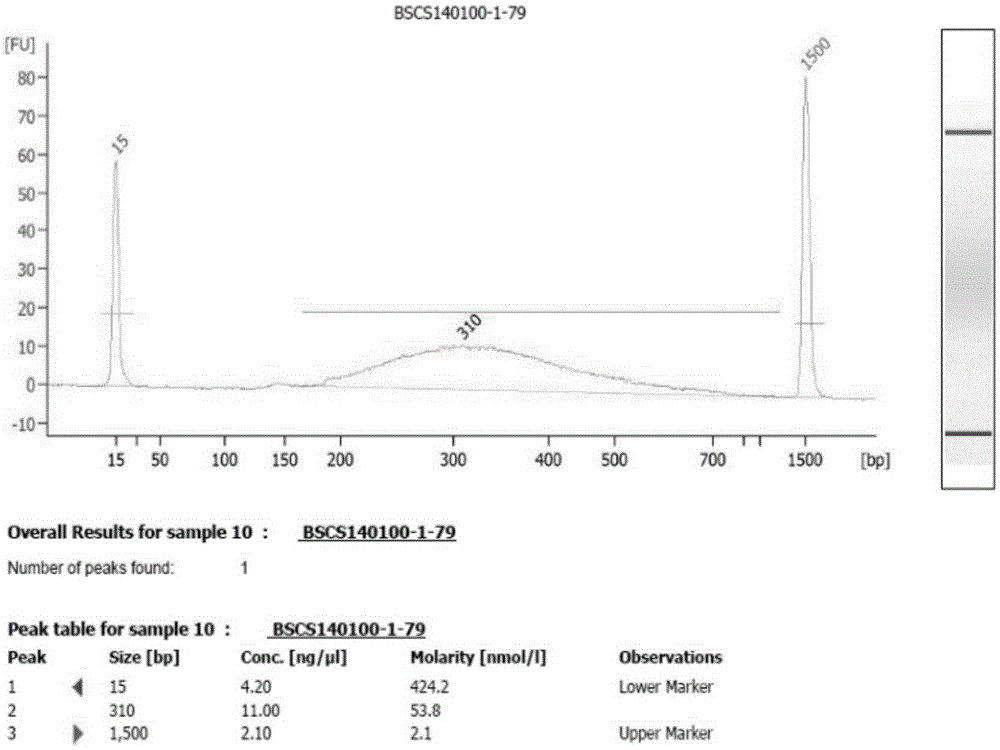
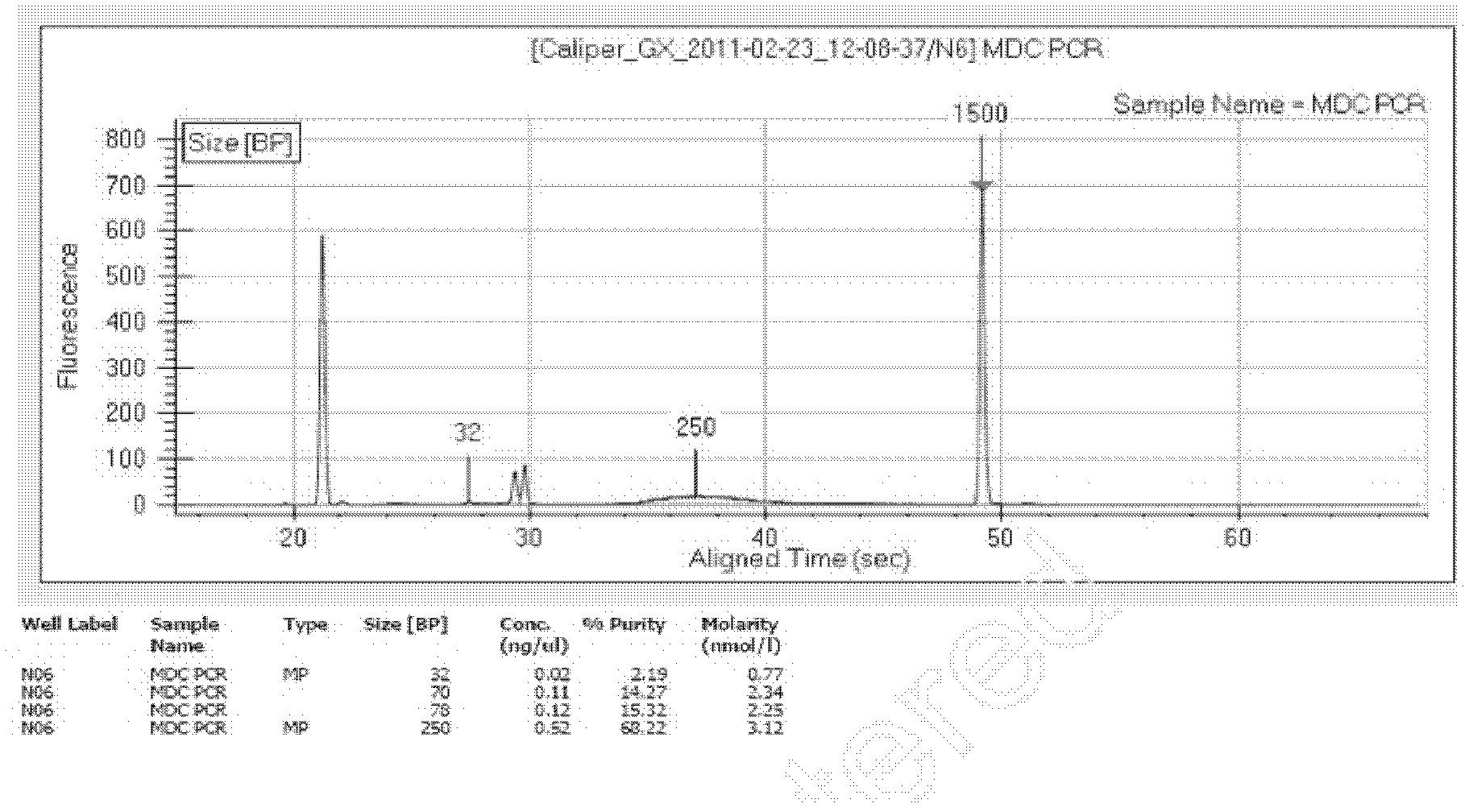
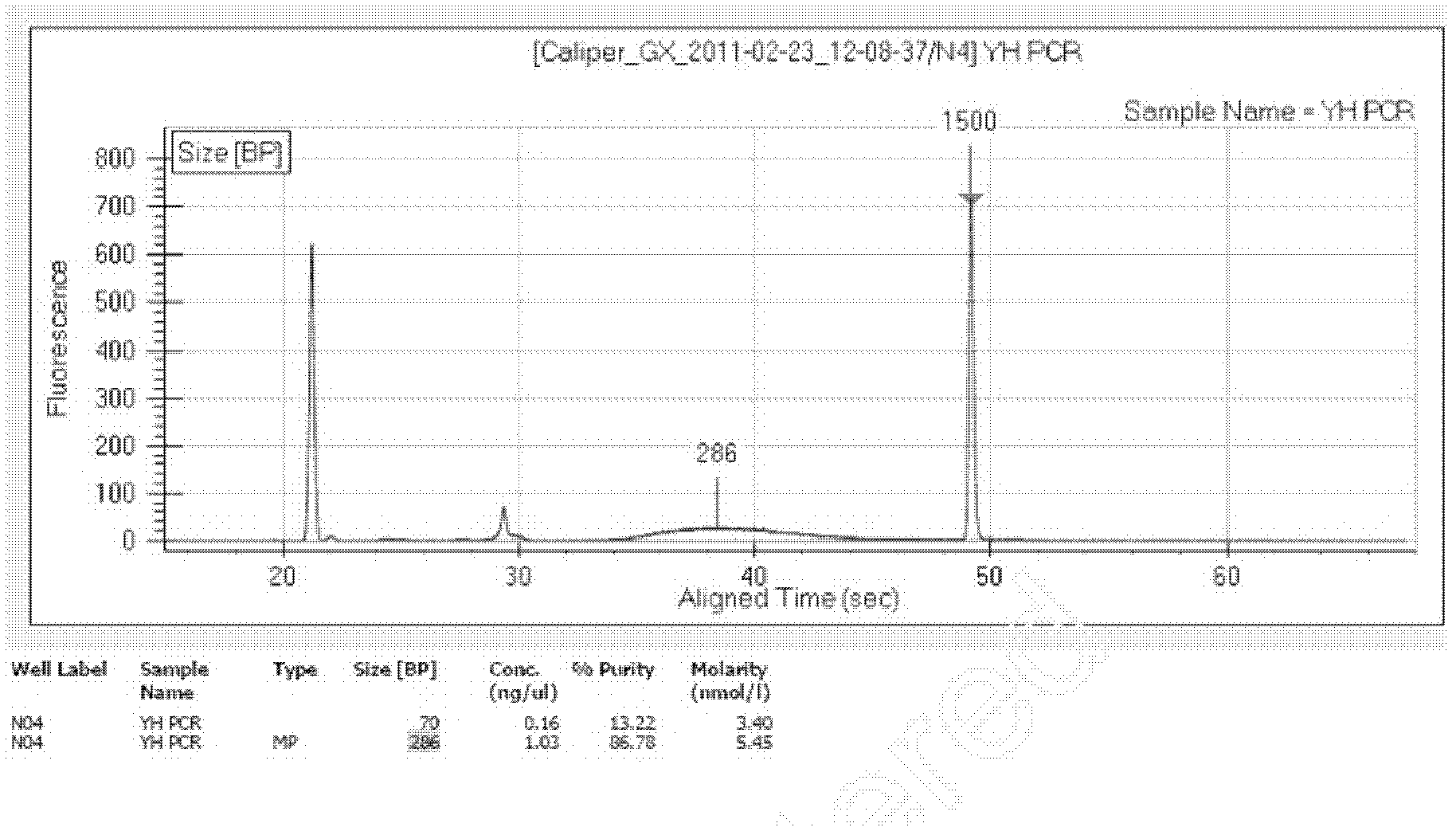
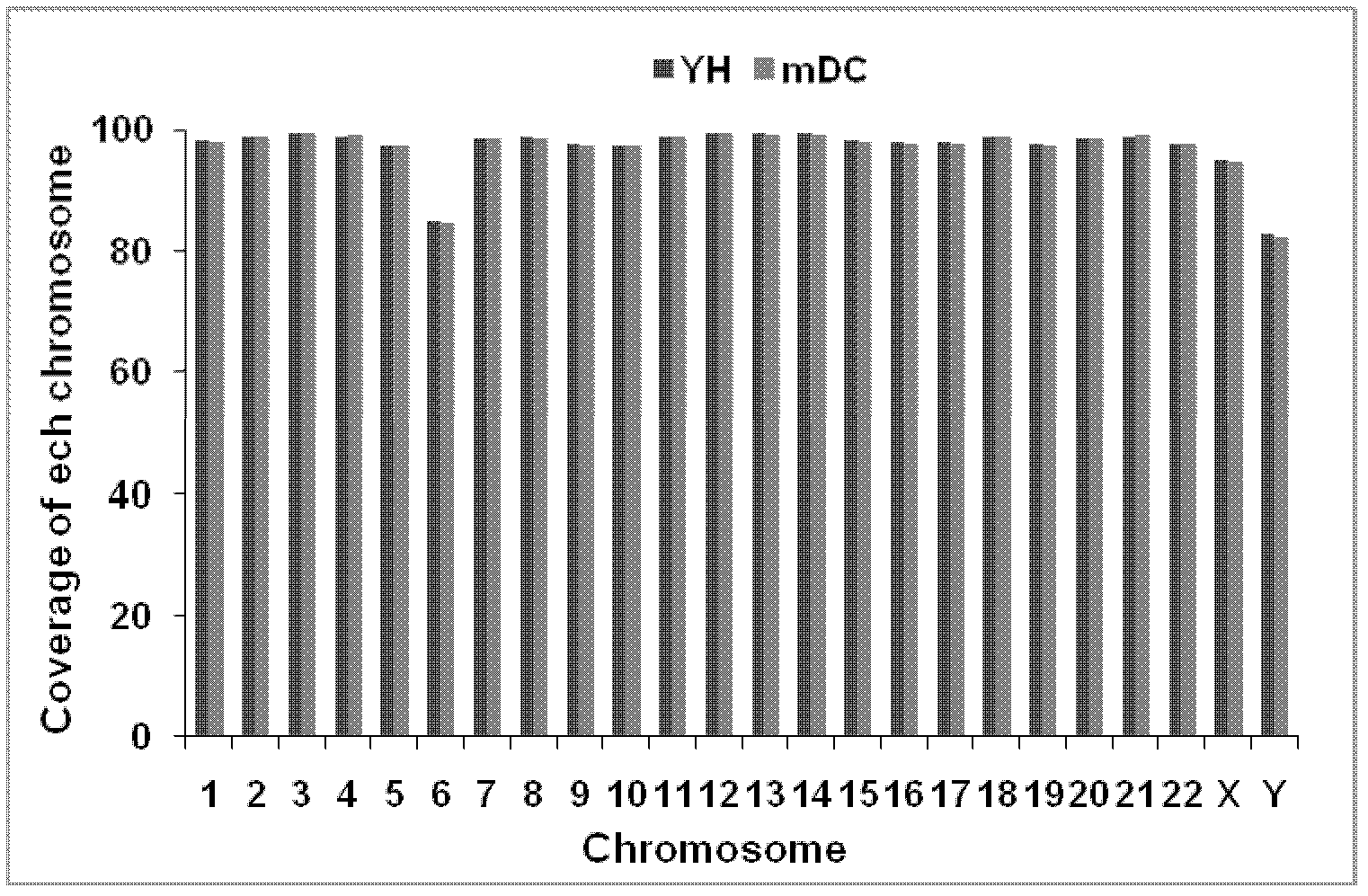
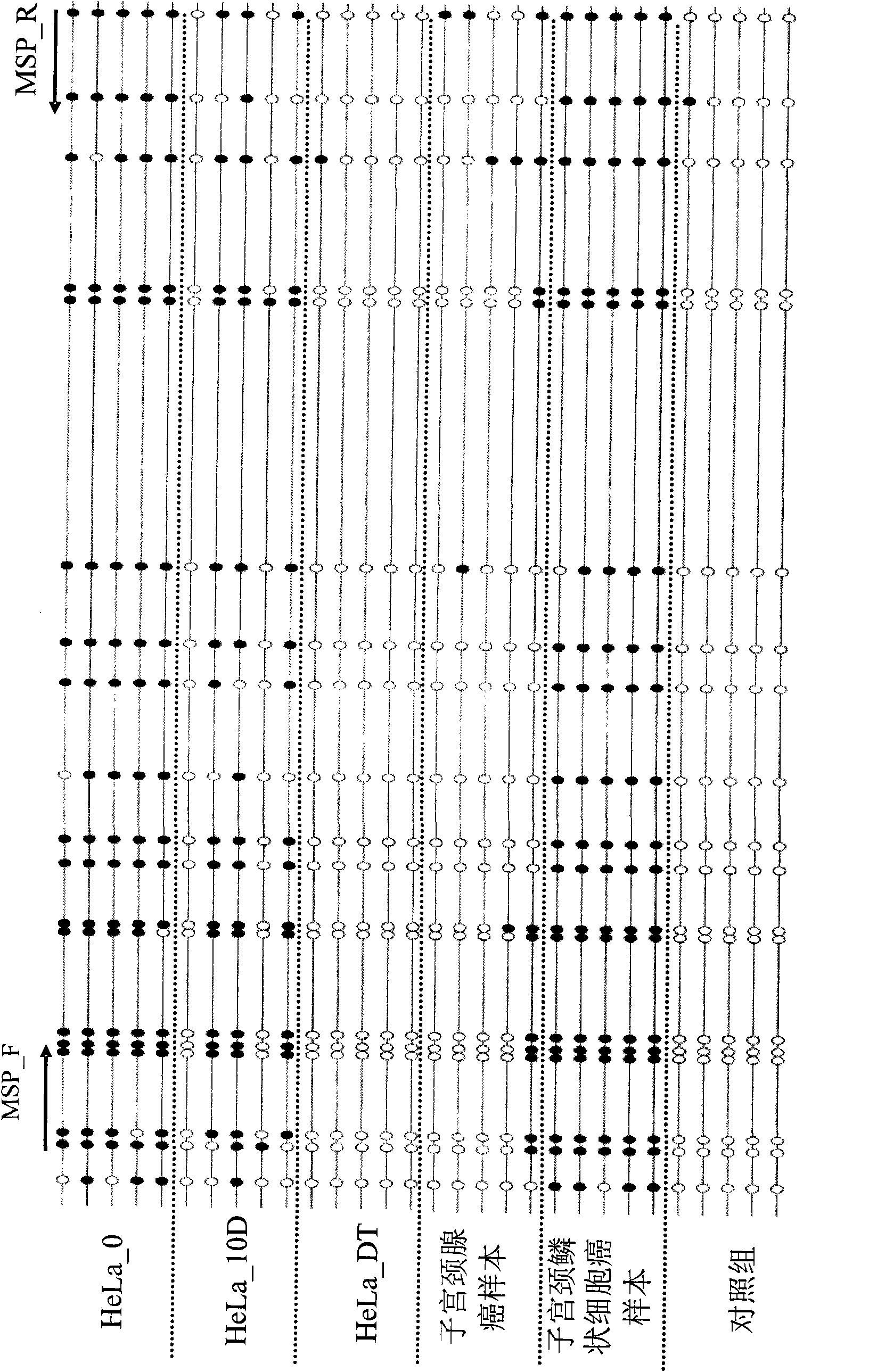
Patents
Literature
38 results about "Bisulfite sequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Bisulfite sequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity.
Cancer screening method
InactiveUS20080311570A1Accurate screening resultEasy diagnosisMicrobiological testing/measurementFermentationScreening cancerScreening method
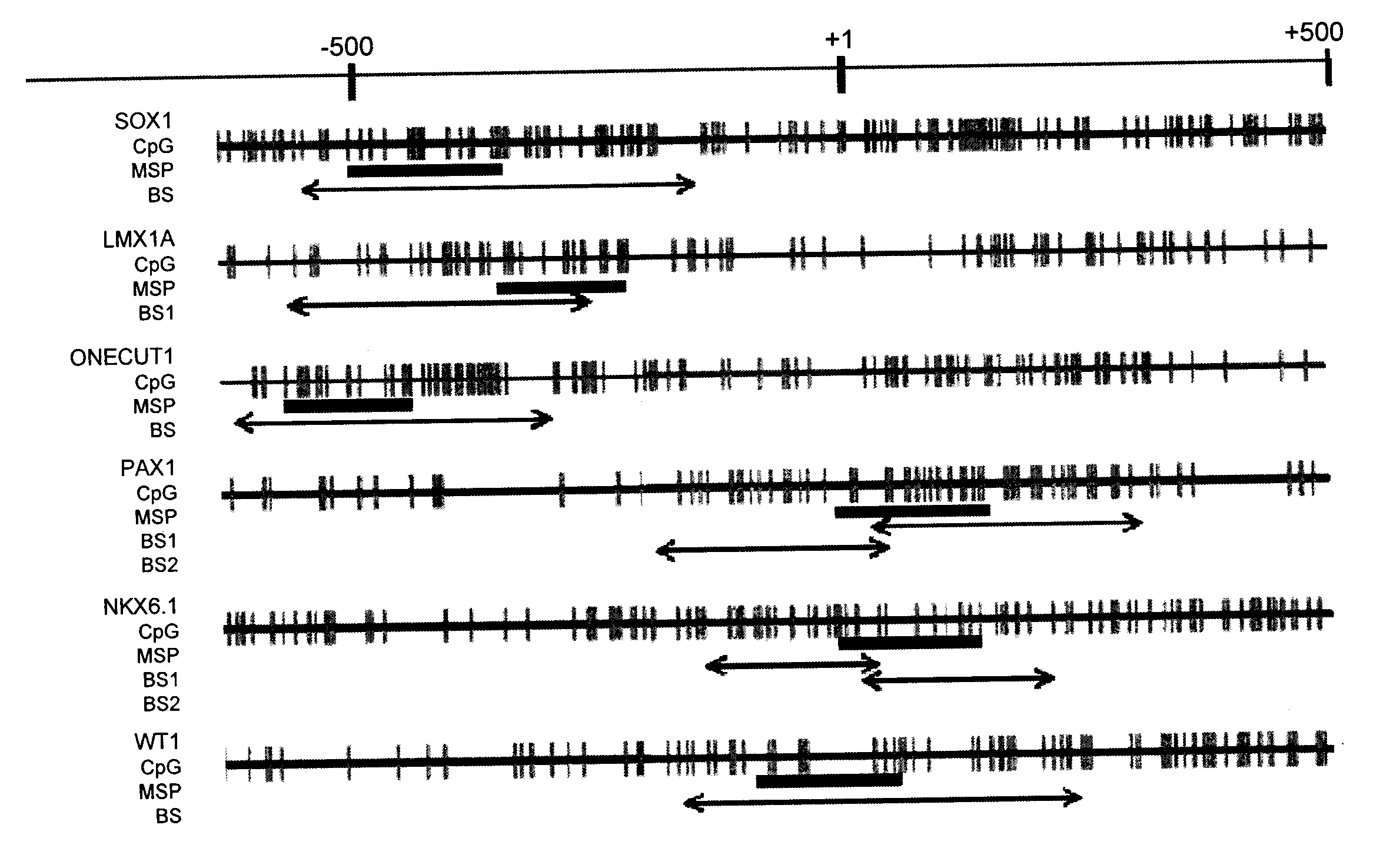
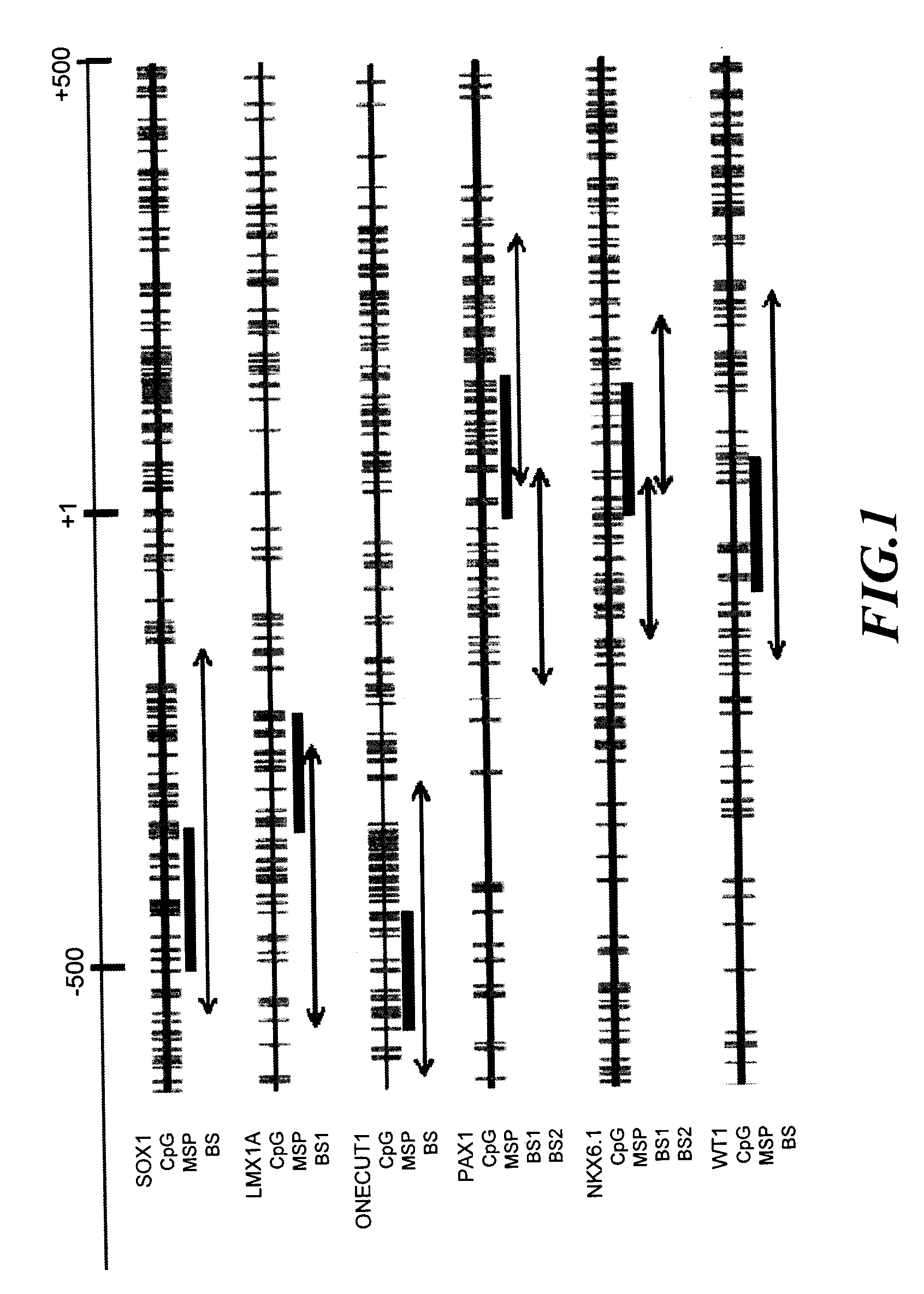
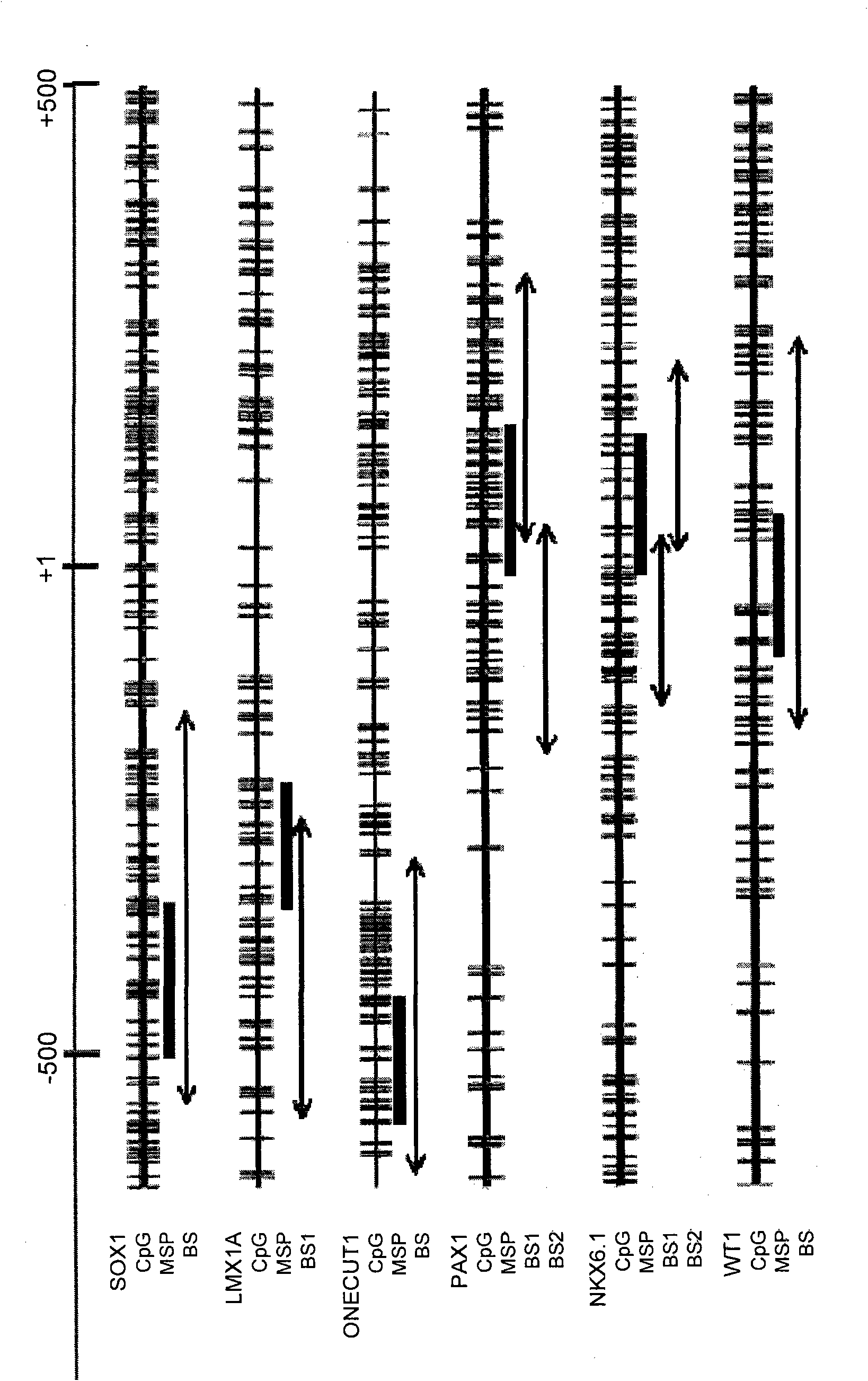
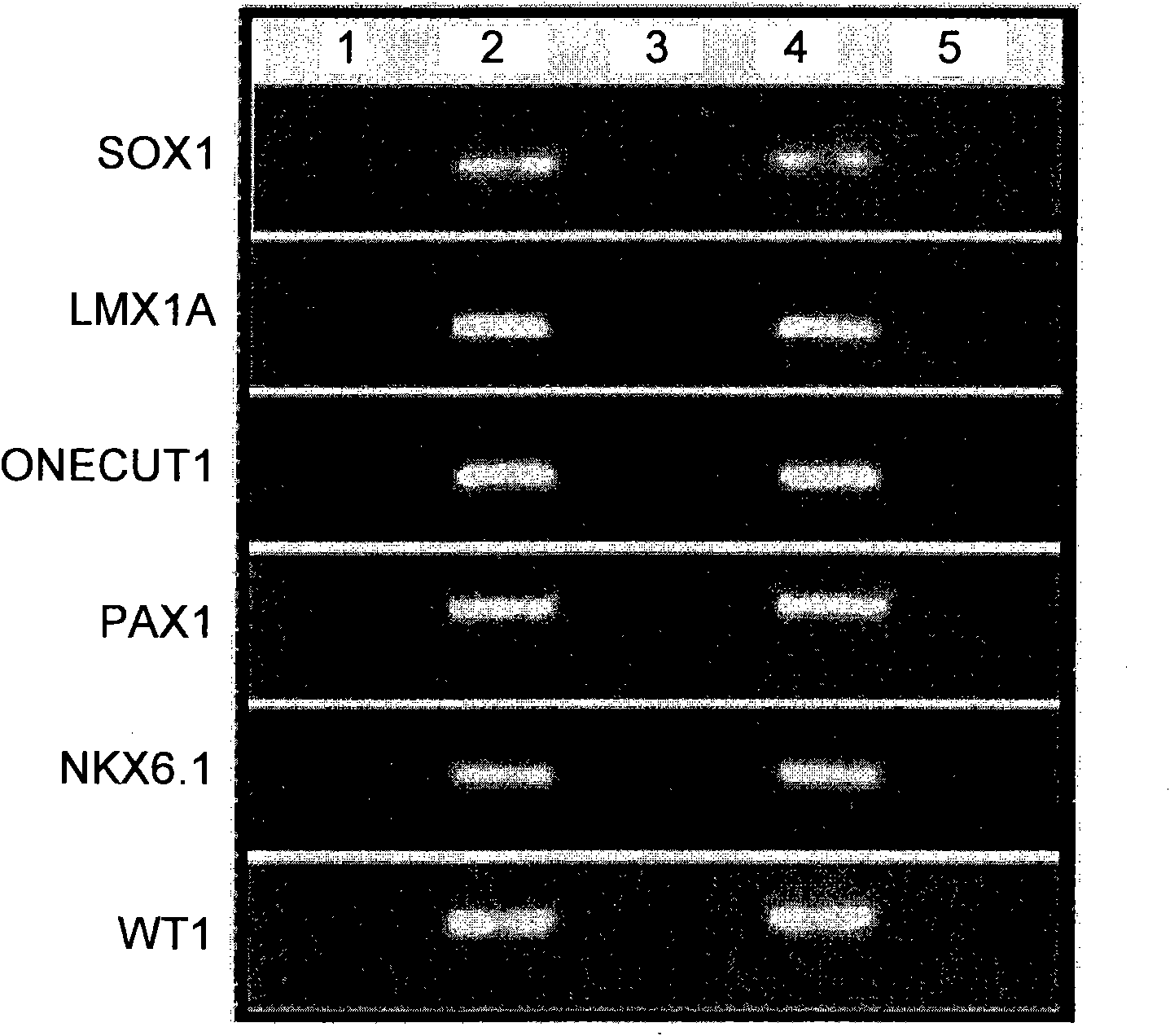
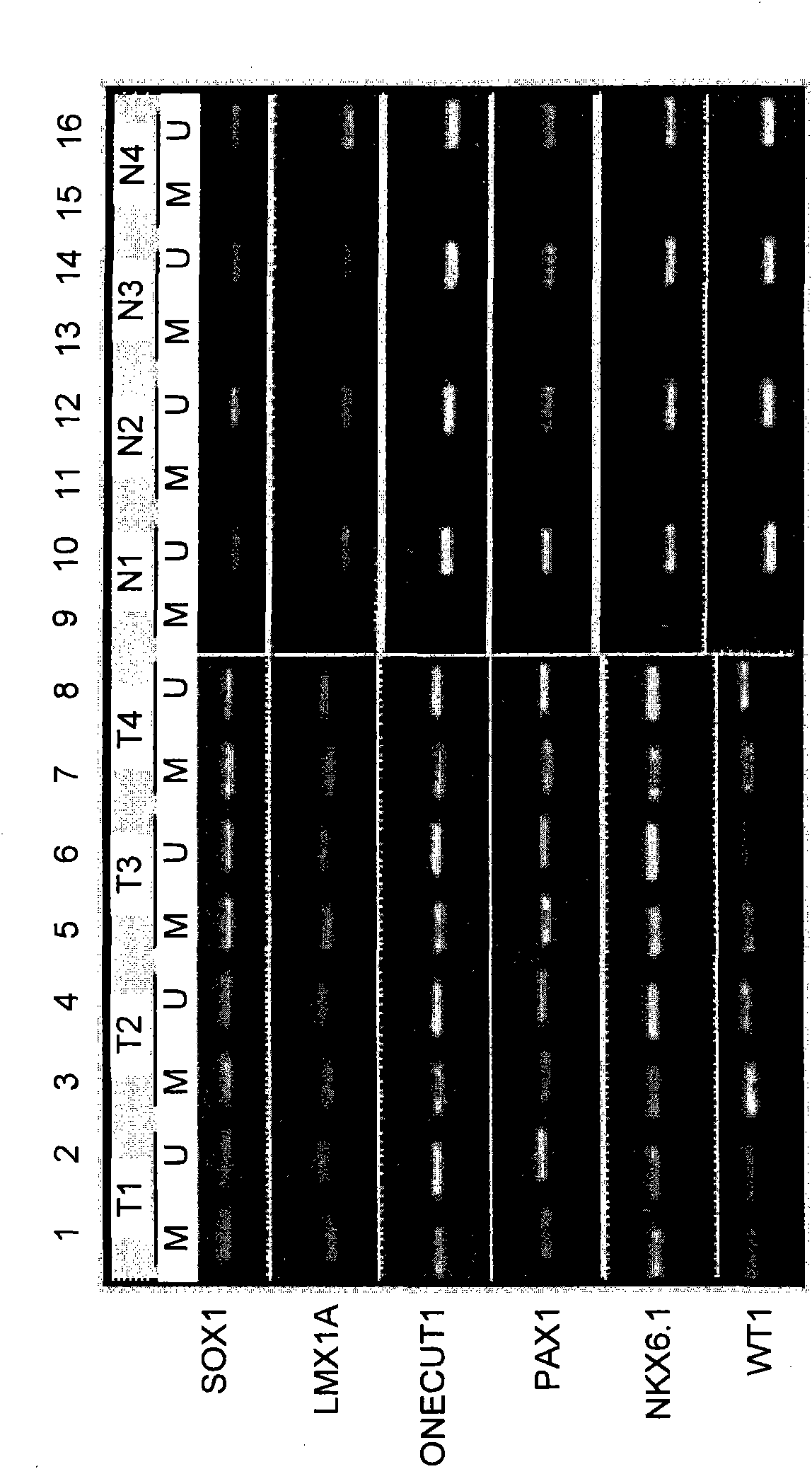
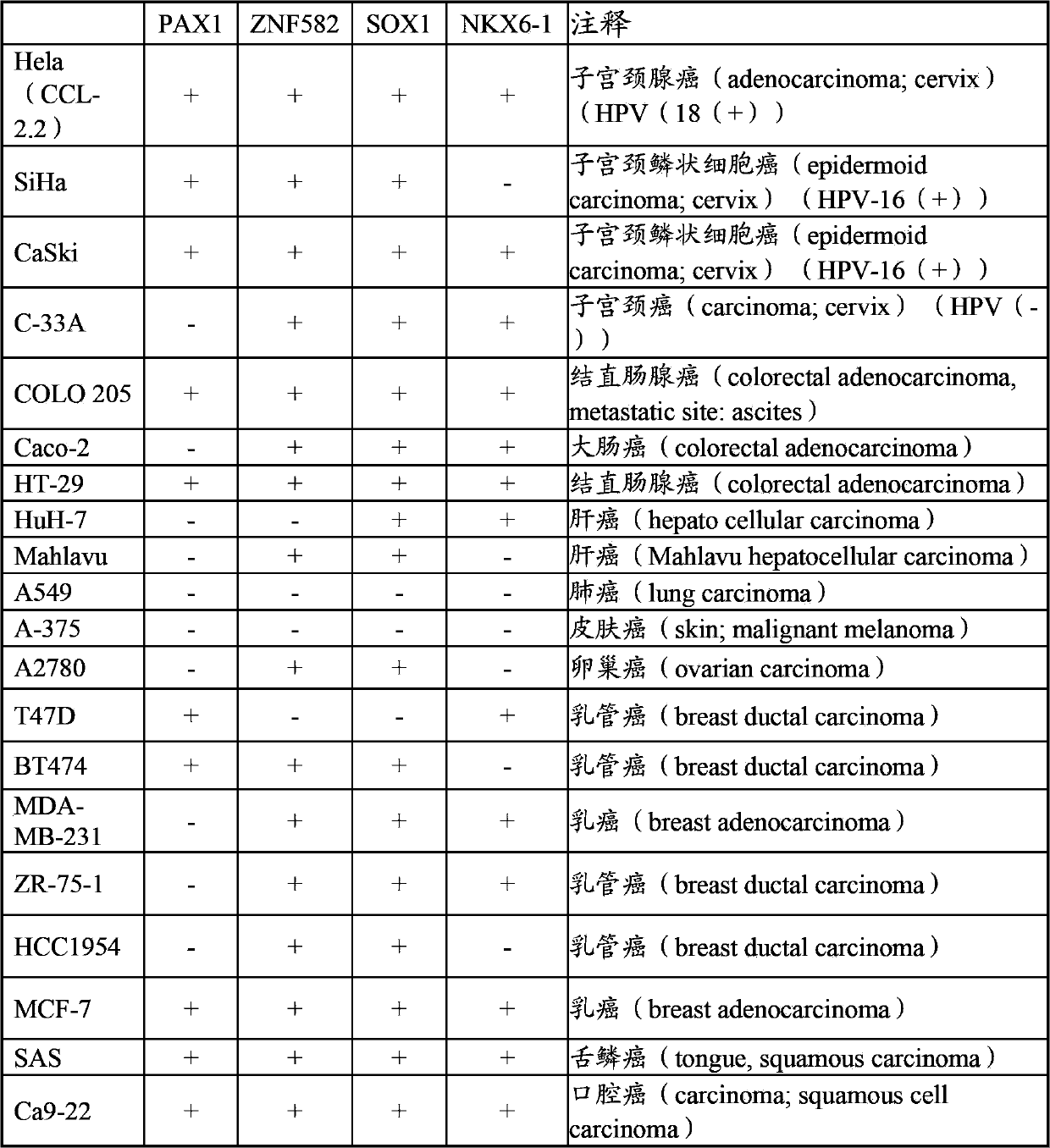
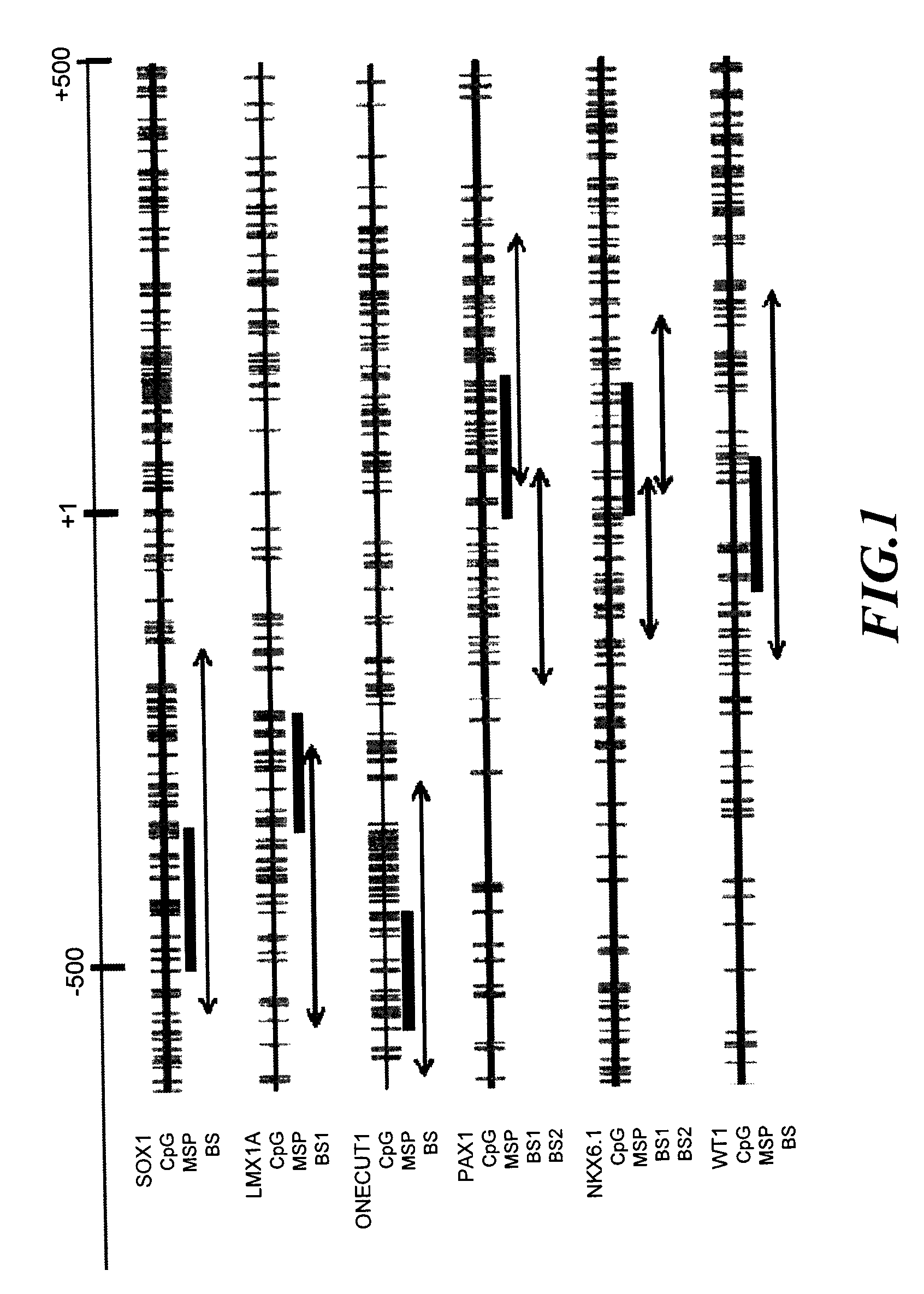
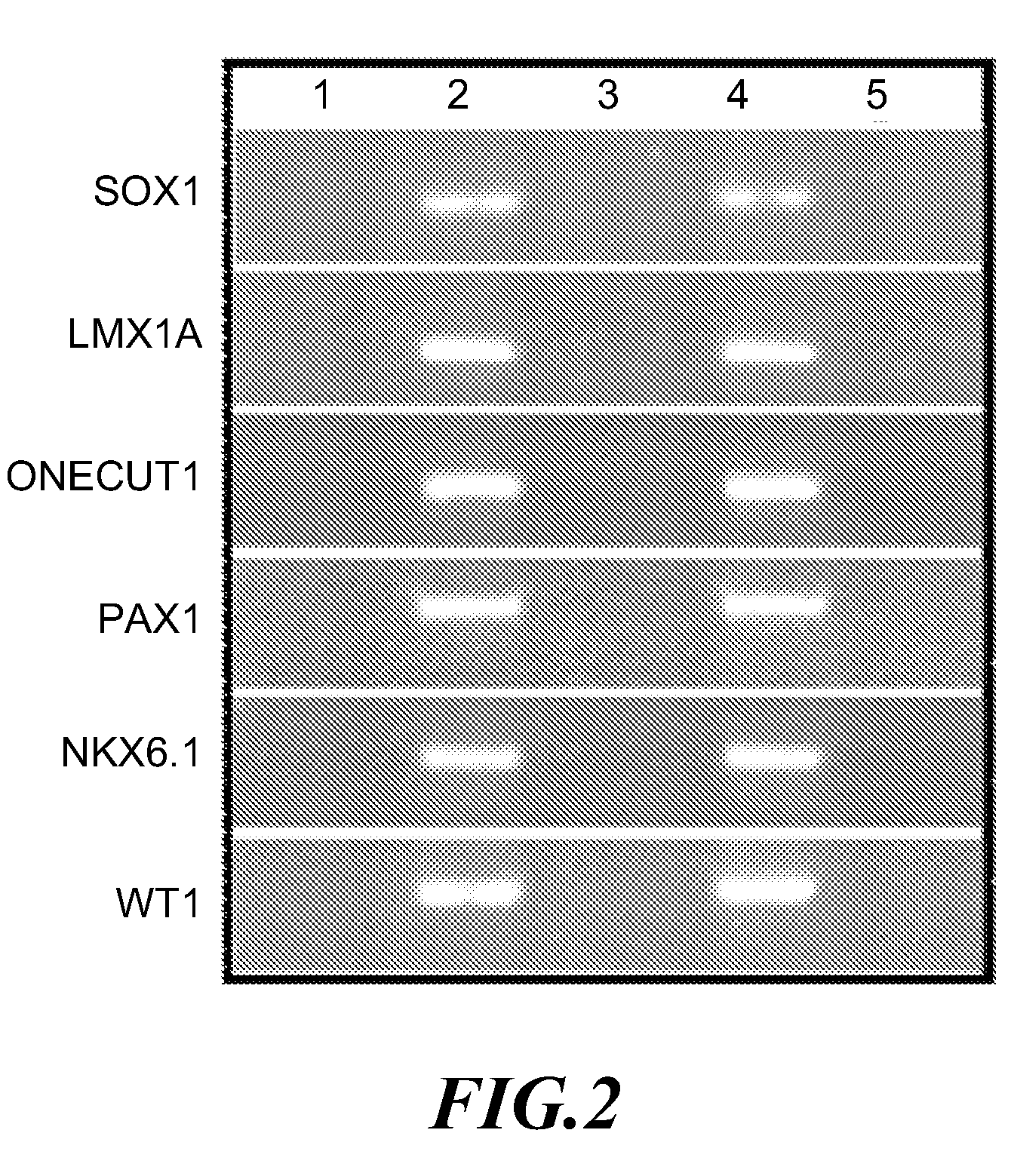
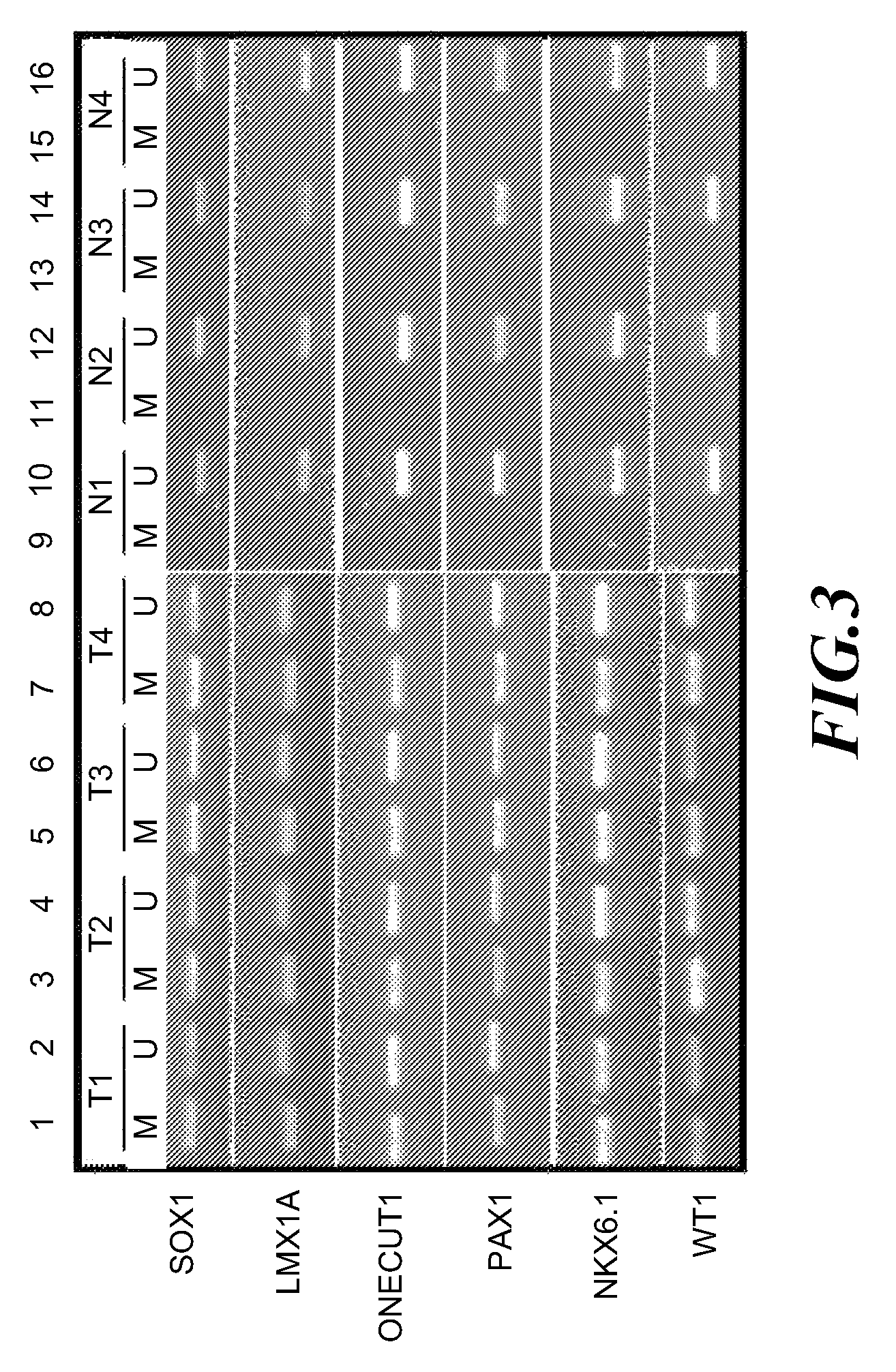
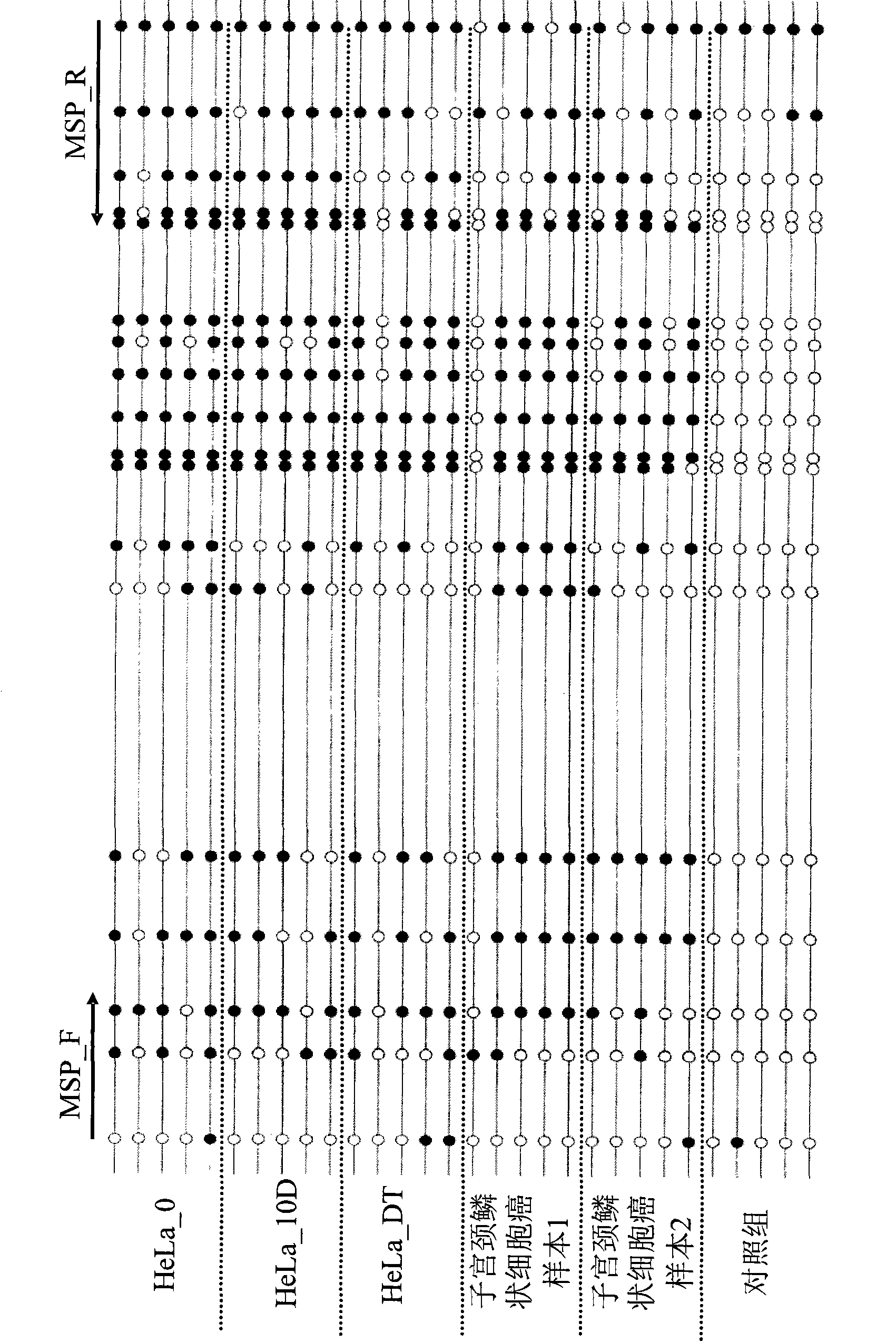
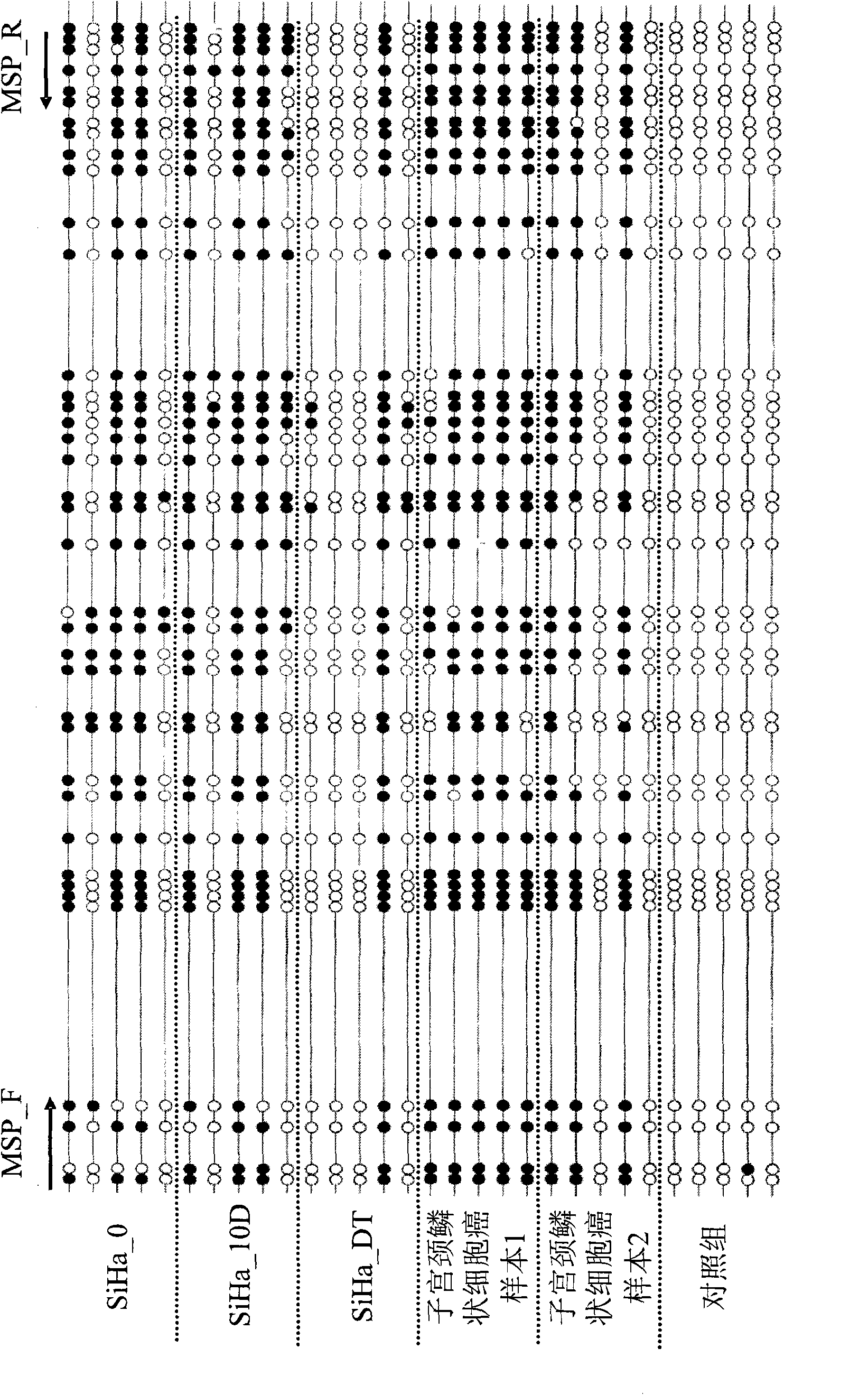
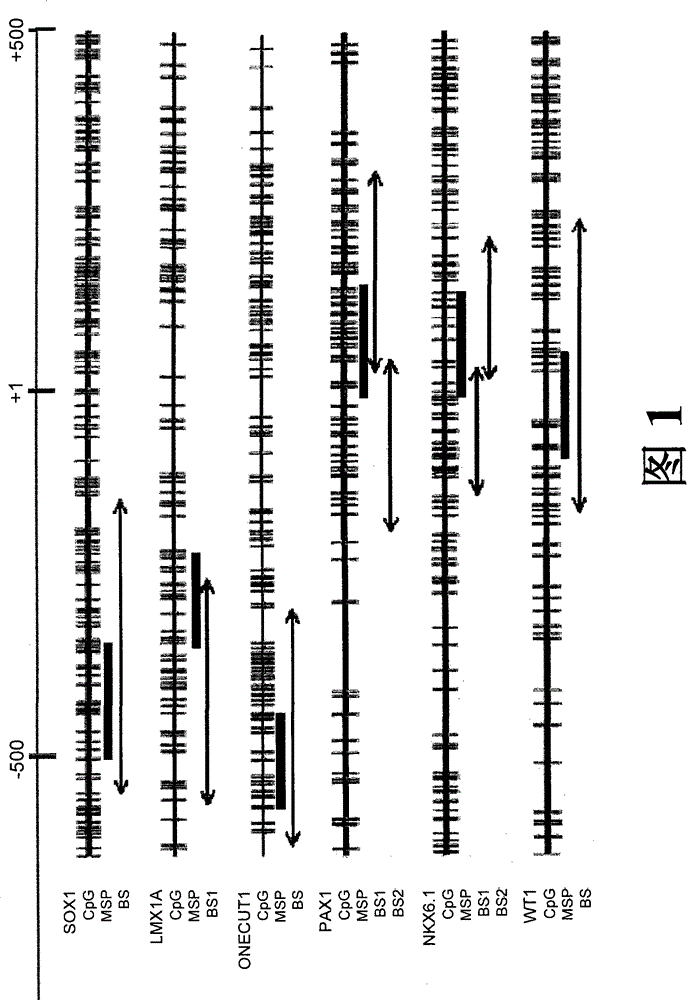
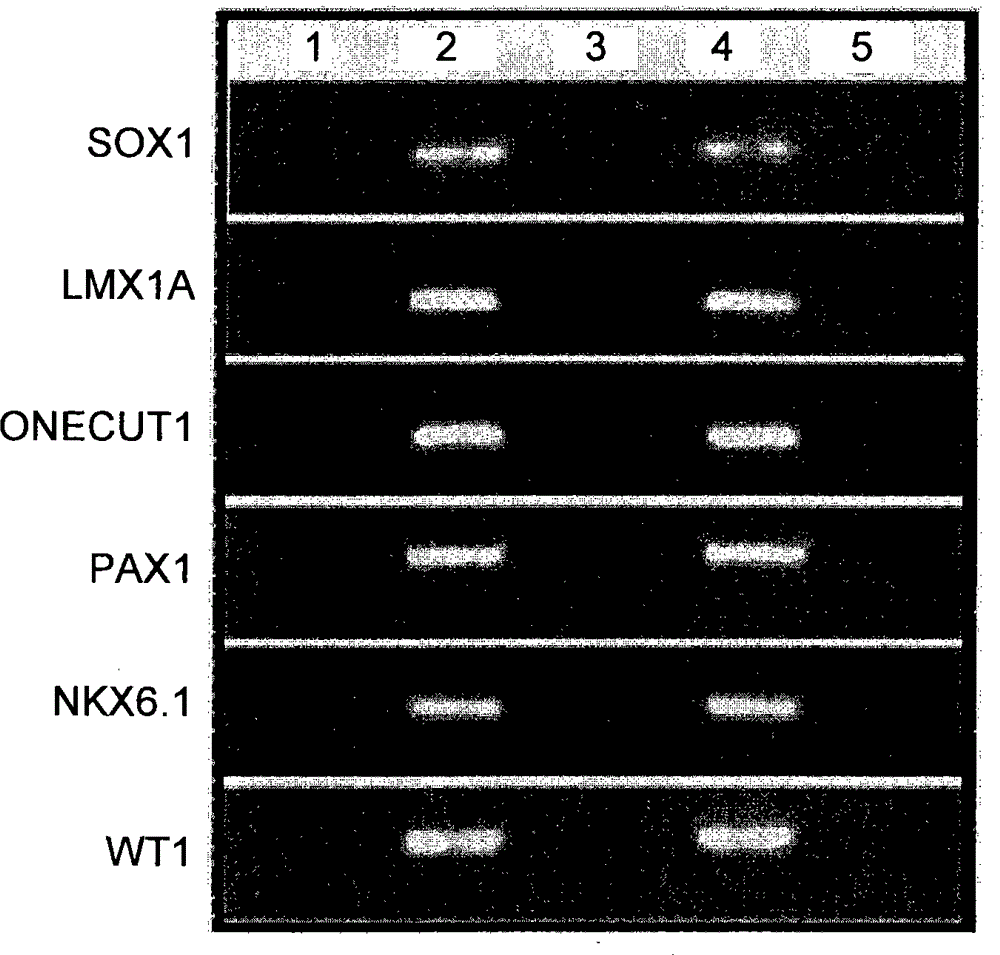
A method for screening cancer comprises the following steps: (1) providing a test specimen; (2) detecting the methylation state of the CpG sequence in at least one target gene within the genomic DNA of the test specimen, wherein the target genes is consisted of SOX1, PAX1, LMX1A, NKX6-1, WT1 and ONECUT1; and (3) determining whether there is cancer or cancerous pathological change in the specimen based on the presence or absence of the methylation state in the target gene; wherein method for detecting methylation state is methylation-specific PCR (MSP), quantitative methylation-specific PCR (QMSP), bisulfite sequencing (BS), microarrays, mass spectrometer, denaturing high-performance liquid chromatography (DHPLC), and pyrosequencing.
Owner:NAT DEFENSE MEDICAL CENT
Sieving and checking method for cancers
ActiveCN101570779AIncreased sensitivityStrong specificityMicrobiological testing/measurementMethylation specific polymeraseSpectrograph
The invention relates to a sieving and checking method for cancers, comprising the following steps: (1), providing a test body to be checked; (2), checking the CpG sequence methylation state of at least one target gene in the genome DNA of the test body to be checked, wherein at least one of the target genes comprises SOX1, PAX1, LMX1A, NKX6-1, WT1 and ONECUT1; and (3), judging whether the test body has cancers or precancerous lesions or not according to the existence or inexistence of the methylation state of the target genes. The methylation state checking method comprises a methylation specific polymerase chain reaction, a quantitative methylation specific polymerase chain reaction, sulphite sequencing, micro-arrays, mass spectrograph analysis or denaturation high-efficiency liquid chromatography, and the like.
Owner:赖鸿政
Whole-genome methylation non-bisulfite sequencing library, construction and applications thereof
InactiveCN110820050ASolve the defect of low usage rateReduce usageNucleotide librariesMicrobiological testing/measurementDihydrouracilPhosphoric acid
The invention relates to the technical field of bioinformatics, particularly to a whole-genome methylation non-bisulfite sequencing library, a construction and applications thereof, wherein the sequencing library comprises a TET enzyme reaction solution, the TET enzyme reaction solution comprises the following independently packaged components: a TET enzyme oxidation buffer solution, and the TET enzyme oxidation buffer solution comprises, by micromole, (20-167)*10<3> parts of HEPES or Tris-Cl, (100-333)*10<3> parts of NaCl, 3.3*10<3> parts of alpha-KG or 2-oxoglutarate, 6.67*10<3> parts of ascorbic acid and 4*10<3> parts of adenosine triphosphate. According to the invention, by combining the kit, Fe(NH4)2(SO4)2 and TET enzyme, 5mc can be oxidized into 5cac, the 5cac is reduced into dihydrouracil under the action of a reducing agent, and T is identified through PCR sequencing, so that the the DNA methylation C-to-T conversion under the non-bisulfite condition is achieved, the defects ofbase imbalance and low sequencing data use rate of the existing methylation sequencing library constructed based on the bisulfite conversion are solved; and the formula further has effects of simplecomponents, extremely low TET enzyme use amount and significant cost reducing.
Owner:BEIJING GENEPLUS TECH +1
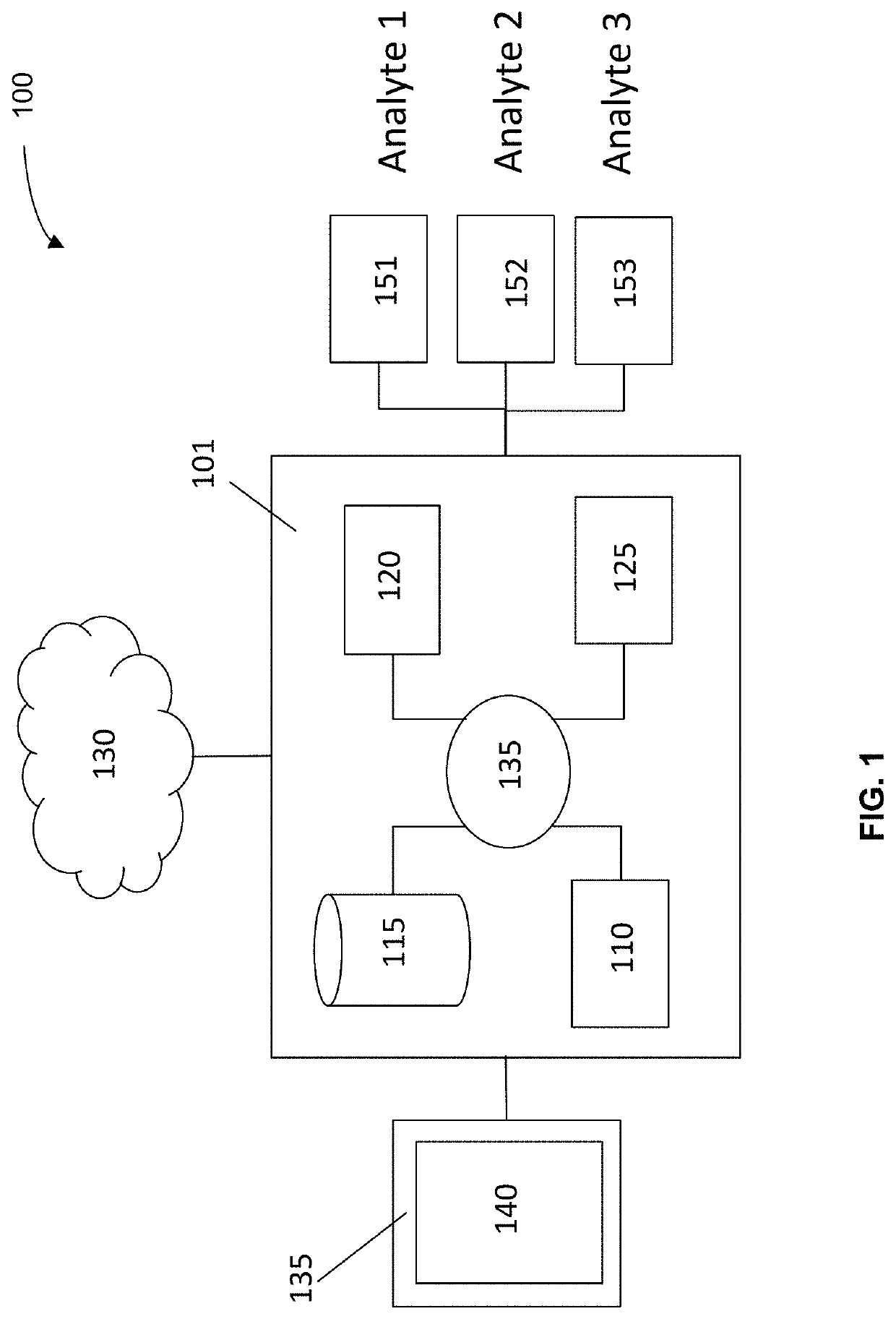
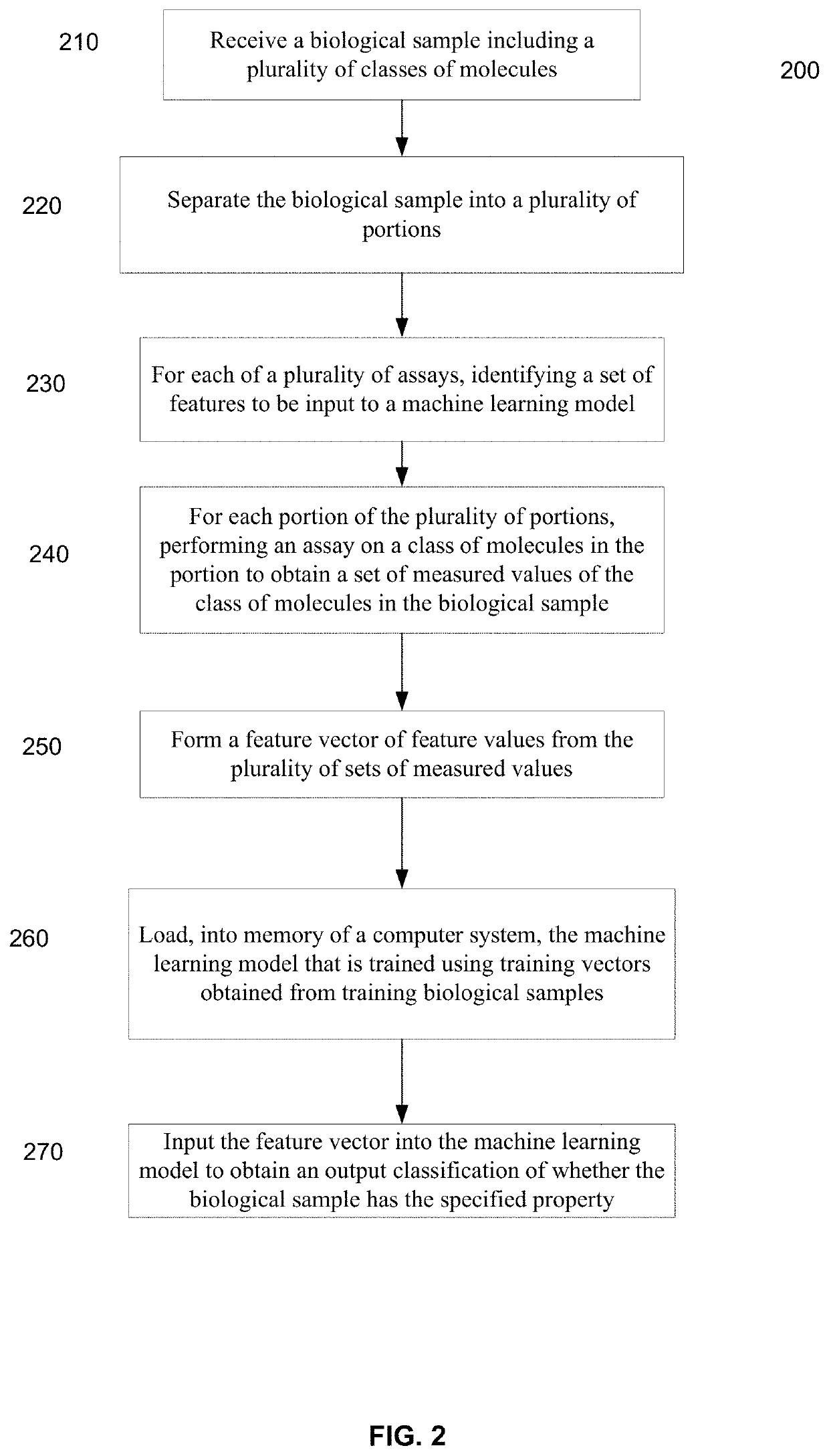
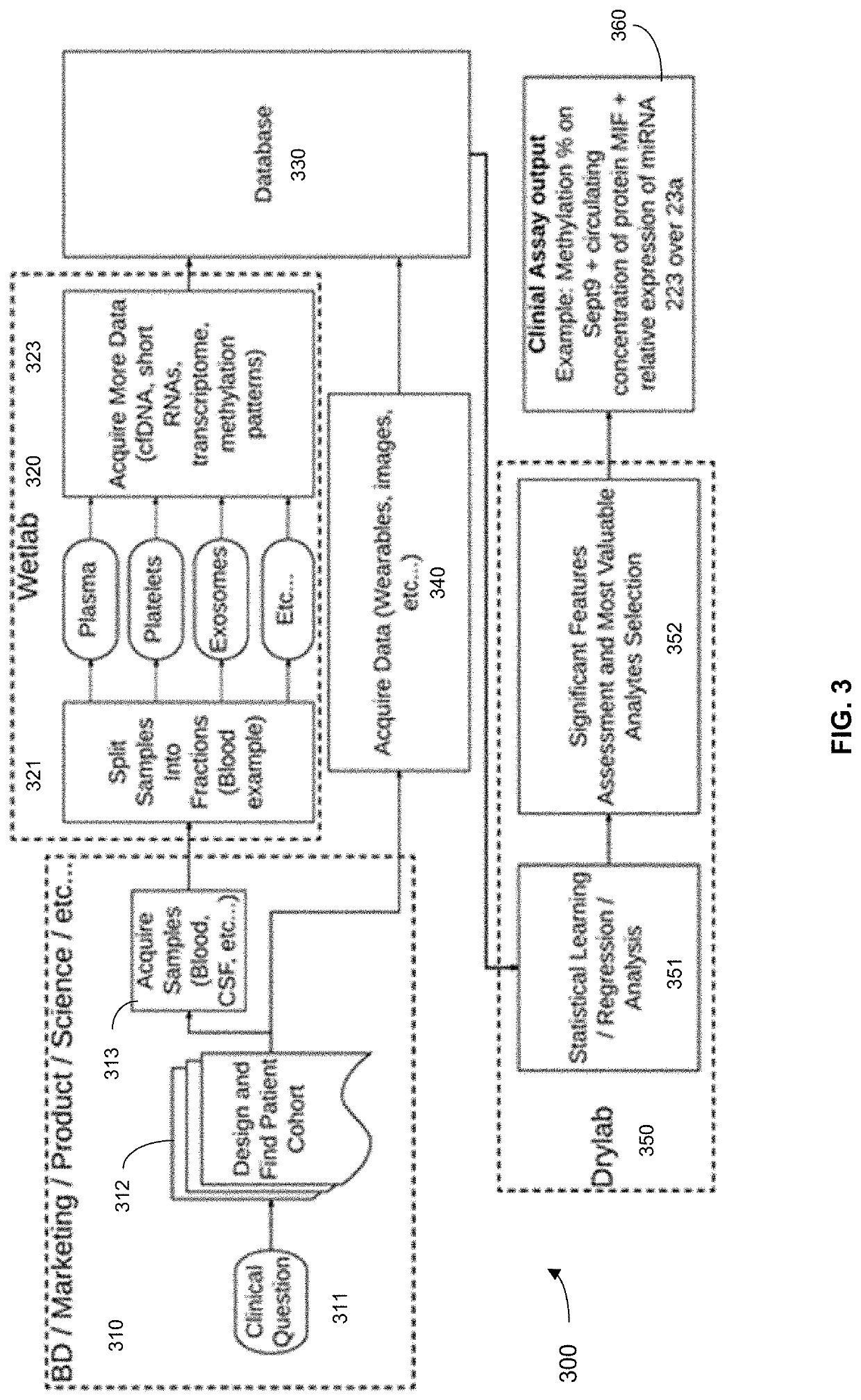
Machine learning implementation for multi-analyte assay development and testing
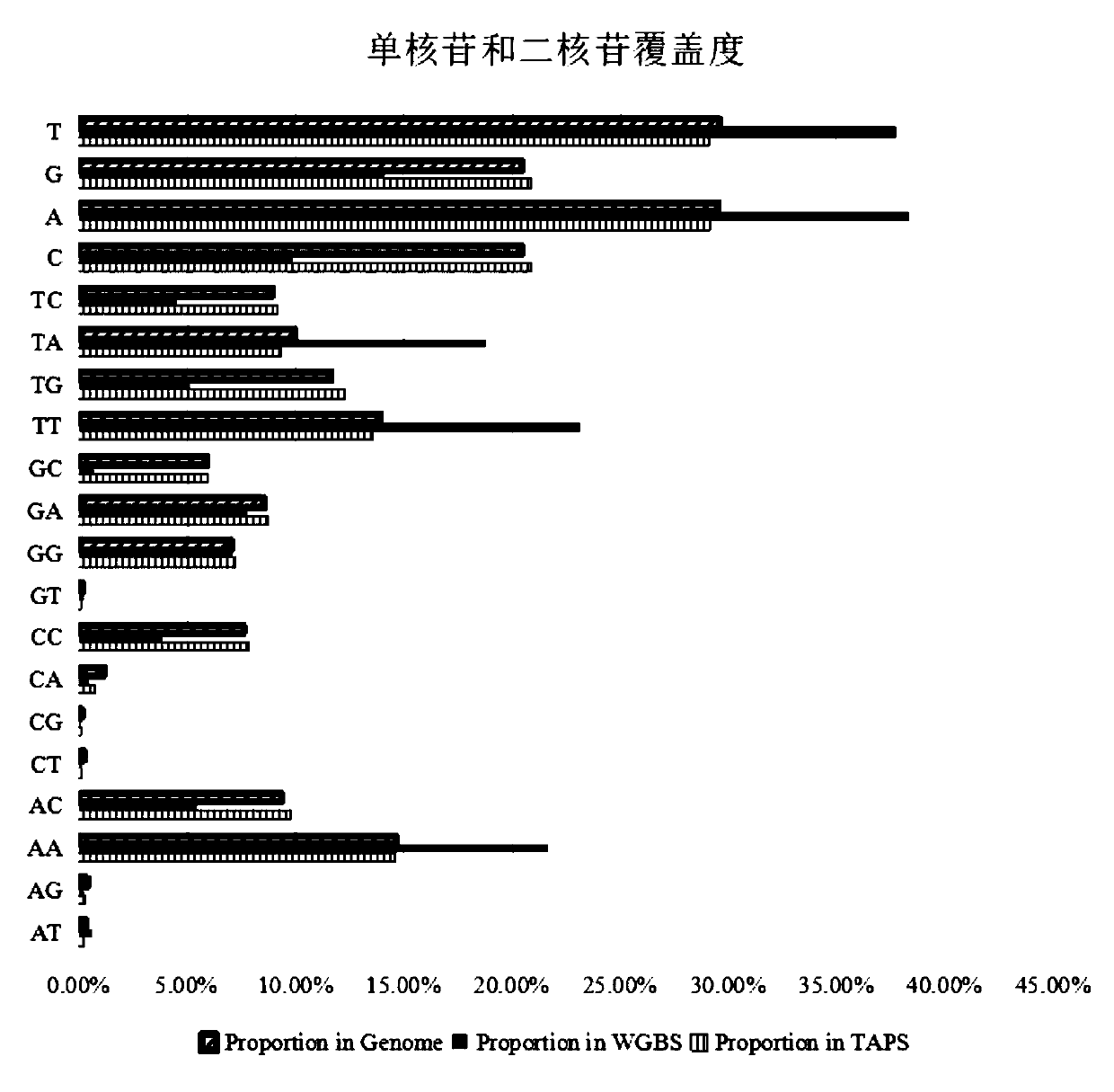
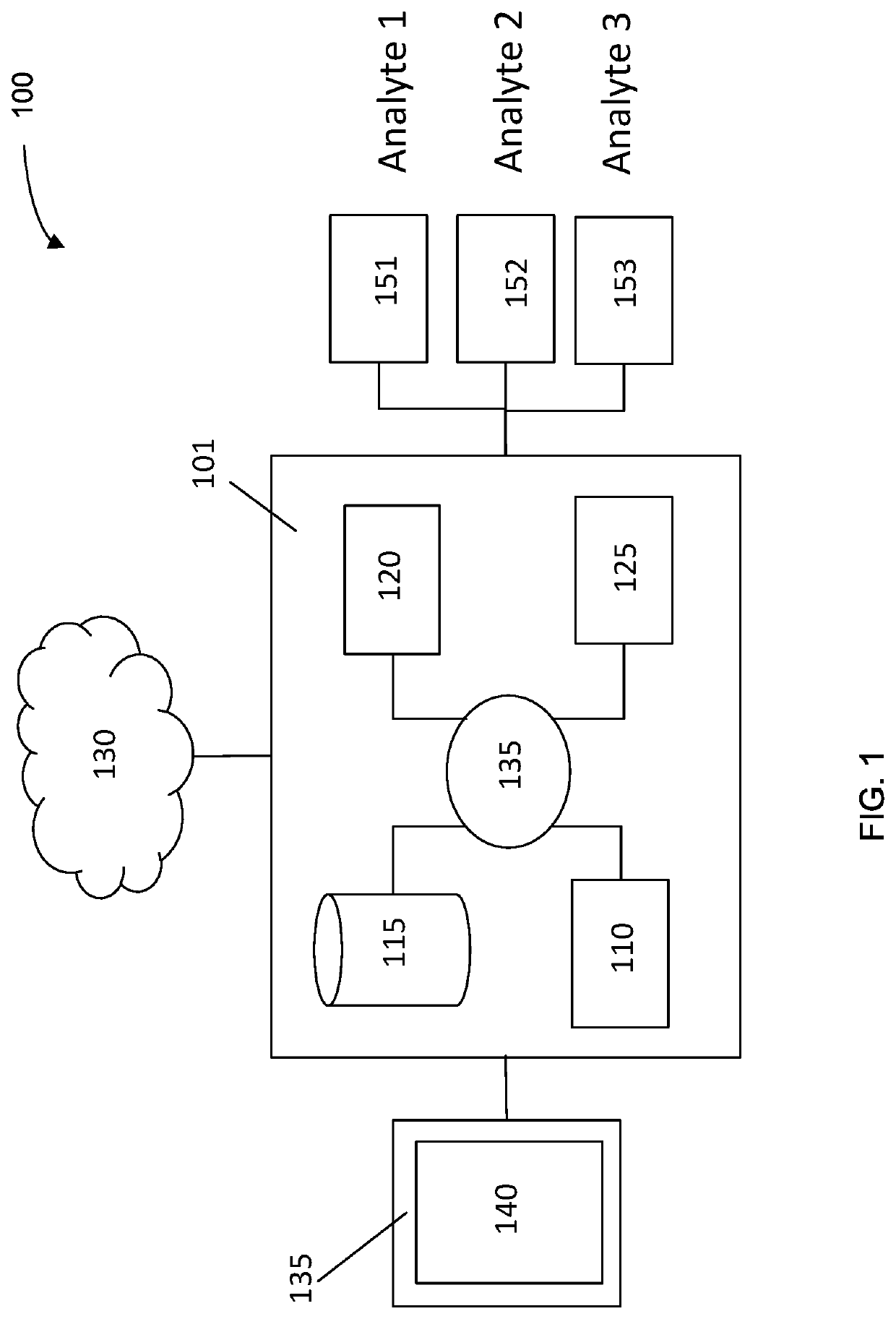
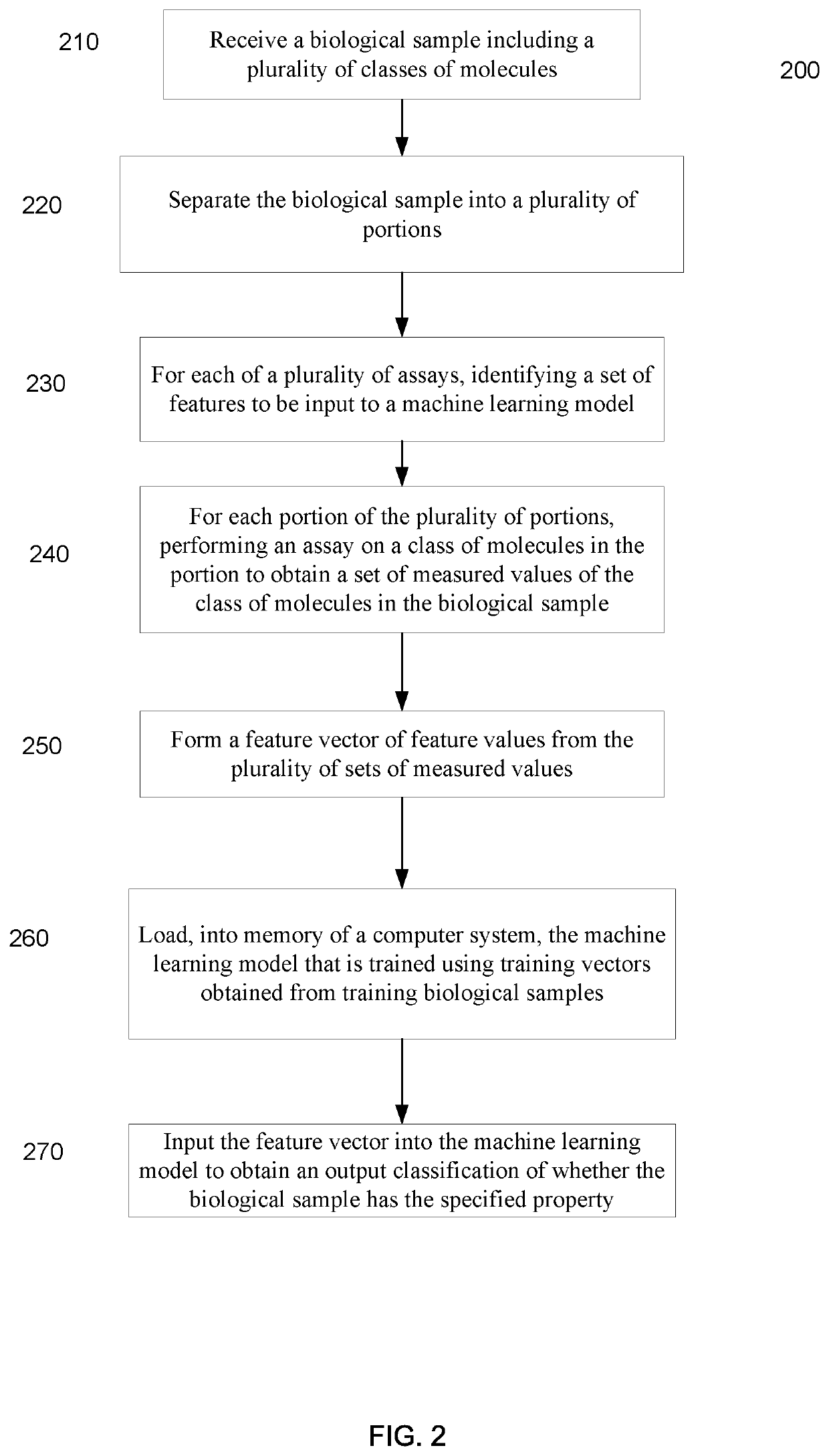
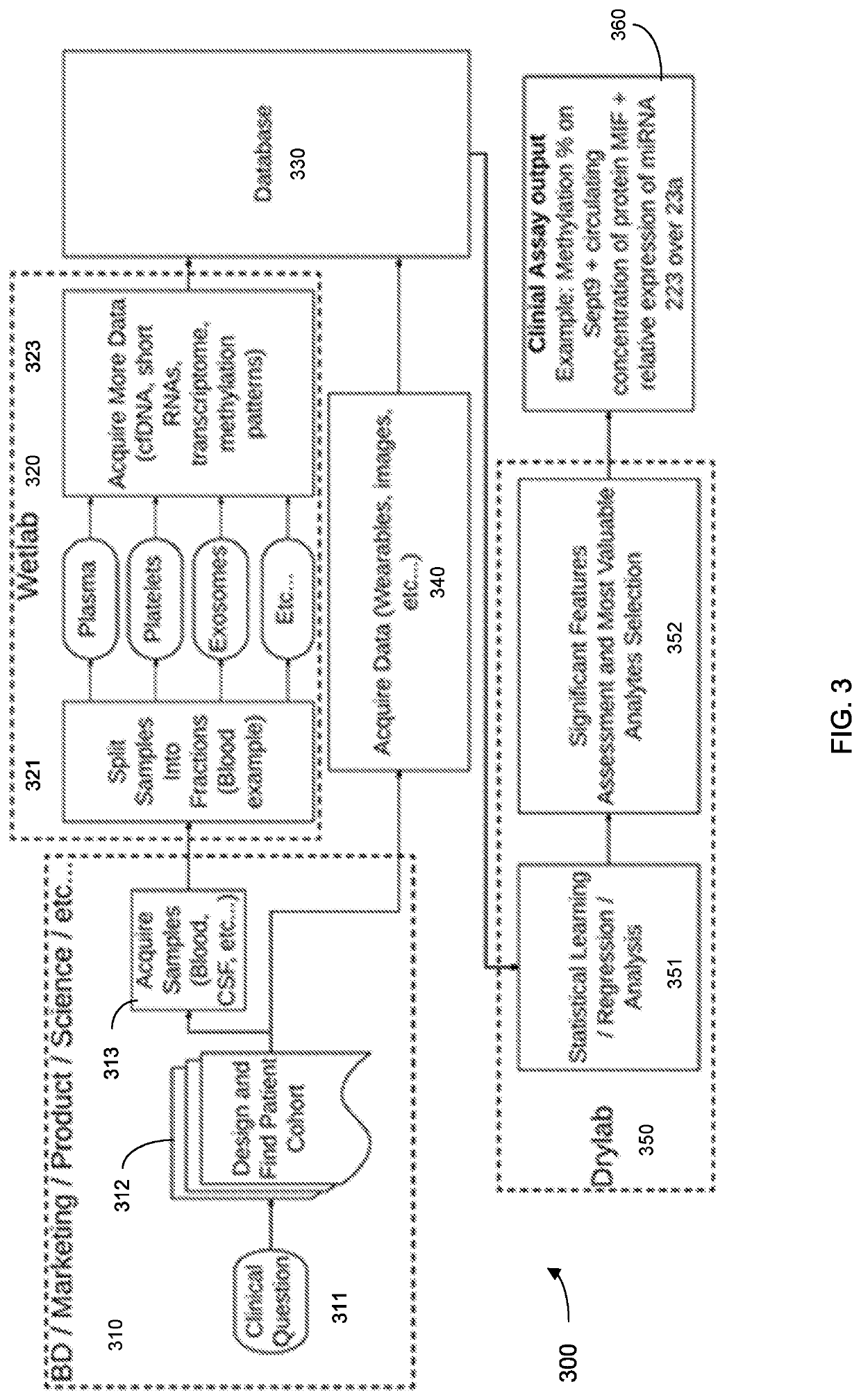
Systems and methods that analyze blood-based cancer diagnostic tests using multiple classes of molecules are described. The system uses machine learning (ML) to analyze multiple analytes, for example cell-free DNA, cell-free microRNA, and circulating proteins, from a biological sample. The system can use multiple assays, e.g., whole-genome sequencing, whole-genome bisulfite sequencing or EM-seq, small-RNA sequencing, and quantitative immunoassay. This can increase the sensitivity and specificity of diagnostics by exploiting independent information between signals. During operation, the system receives a biological sample, and separates a plurality of molecule classes from the sample. For a plurality of assays, the system identifies feature sets to input to a machine learning model. The system performs an assay on each molecule class and forms a feature vector from the measured values. The system inputs the feature vector into the machine learning model and obtains an output classification of whether the sample has a specified property.
Owner:FREENOME HLDG INC
Cancer screening method
ActiveCN101864480AAccurate screening effectMicrobiological testing/measurementMethylation specific polymeraseScreening method
The invention relates to a cancer screening method which comprises the following steps that: (1) providing a tested body; (2) testing the CpG sequence methylation state of at least one target gene in genome DNA of the tested body, and the target gene comprises PTPRR, ZNF582, PDE8B and DBC1; (3) judging whether the tested body has cancer or lesions before cancer according to that whether the methylation state of the at least one target gene exists; and the methylation state detection method is methylation-specific PCR, MSP, quantitative methylation-specific PCR, QMSP, bisulfite sequencing (BS), microarrays, massspectrometer, denaturing high-performance liquidchromatography (DHPLC) and the like.
Owner:NAT DEFENSE MEDICAL CENT
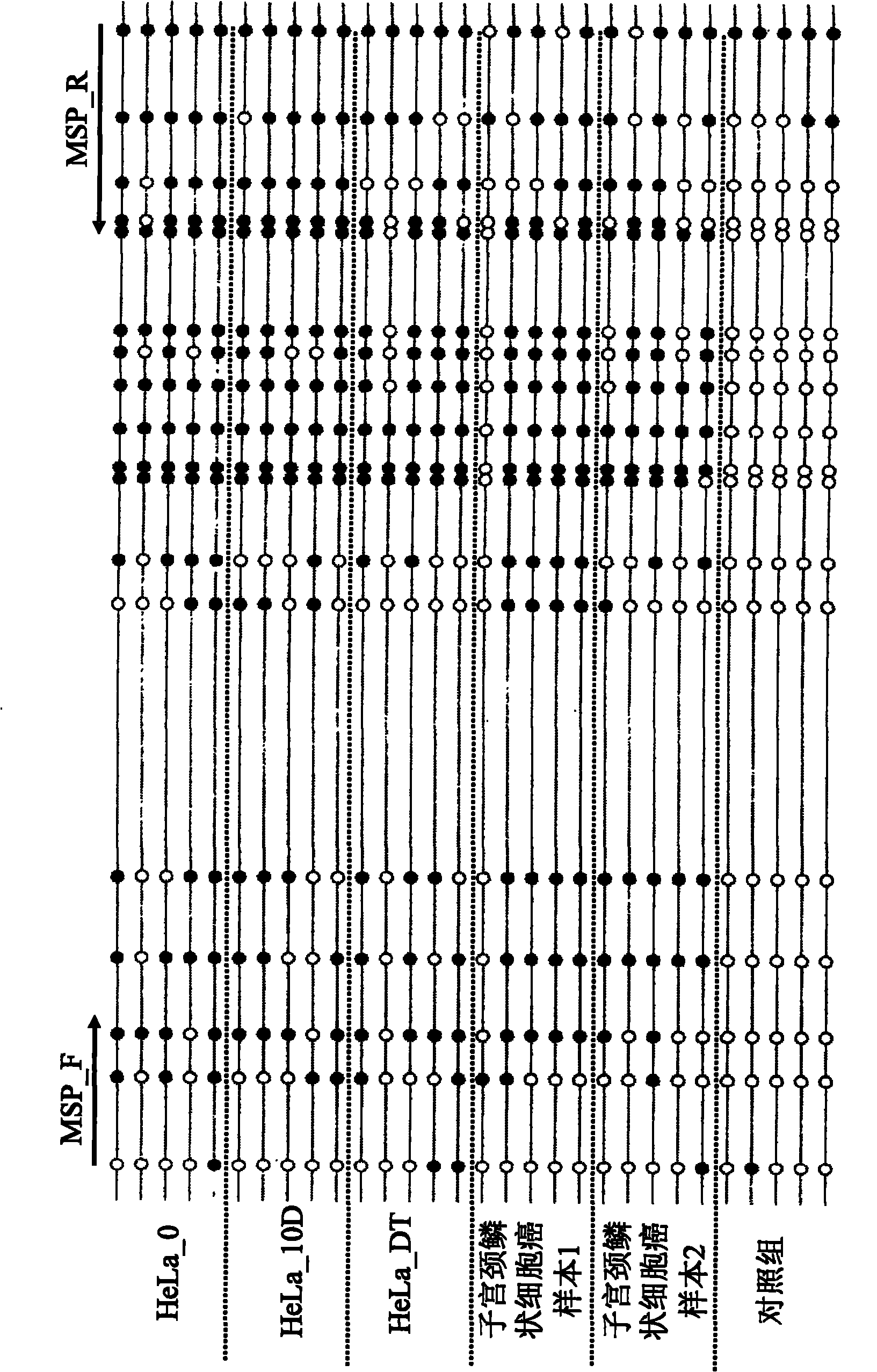
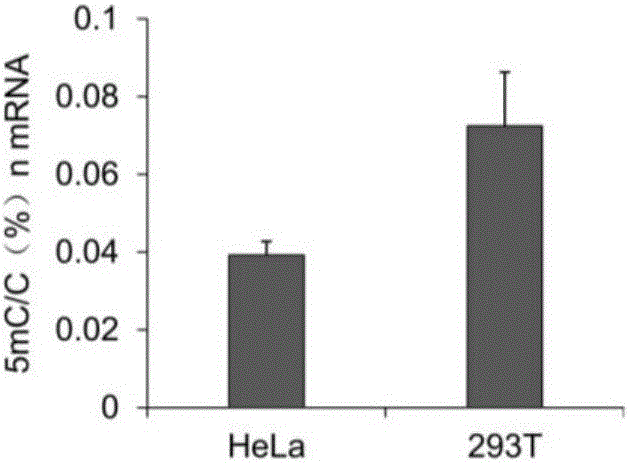
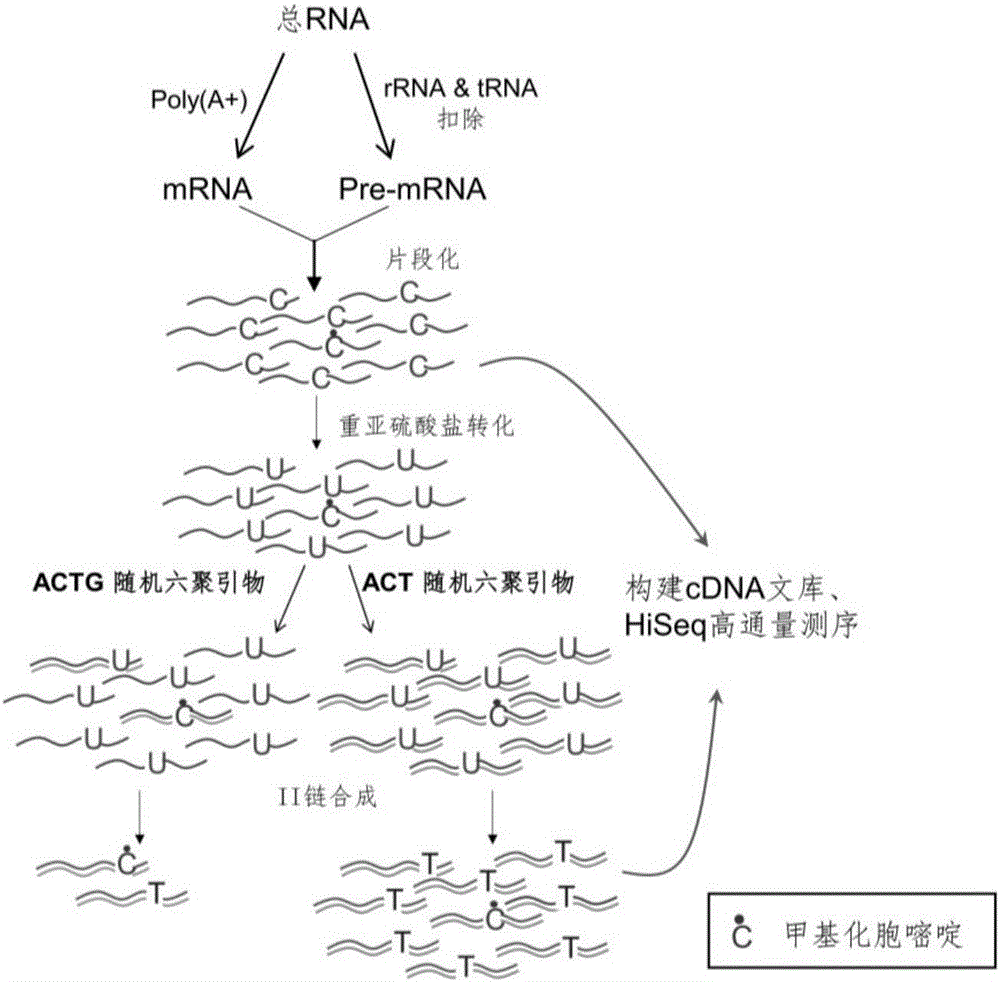
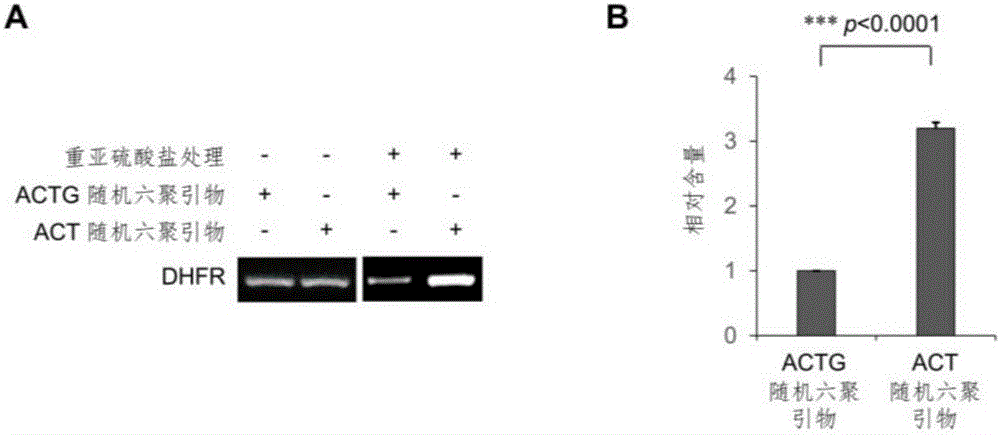
Library construction method for RNA 5mC bisulfite sequencing and application of library
ActiveCN105132409AEfficient matchingHigh amplification efficiencyMicrobiological testing/measurementLibrary creationBisulfite sequencingRandom hexamer
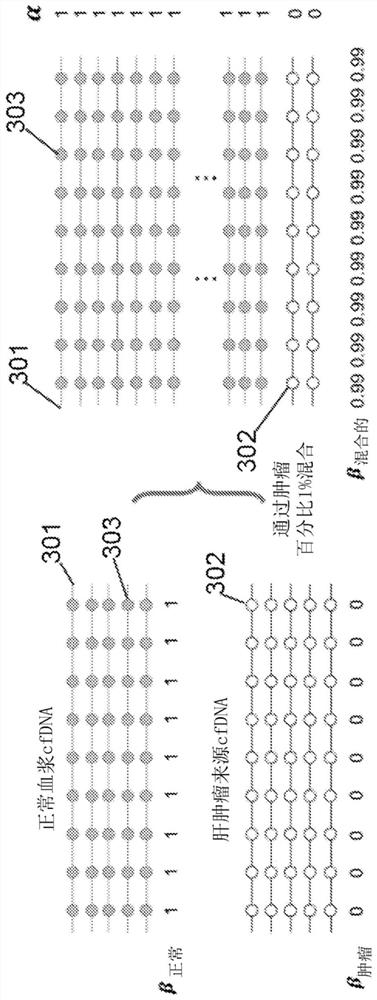
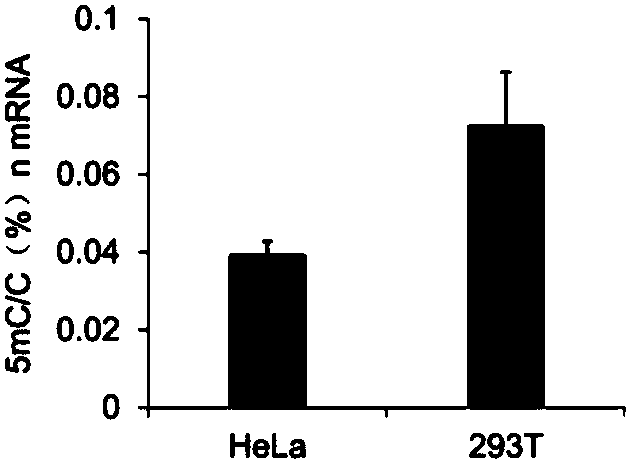
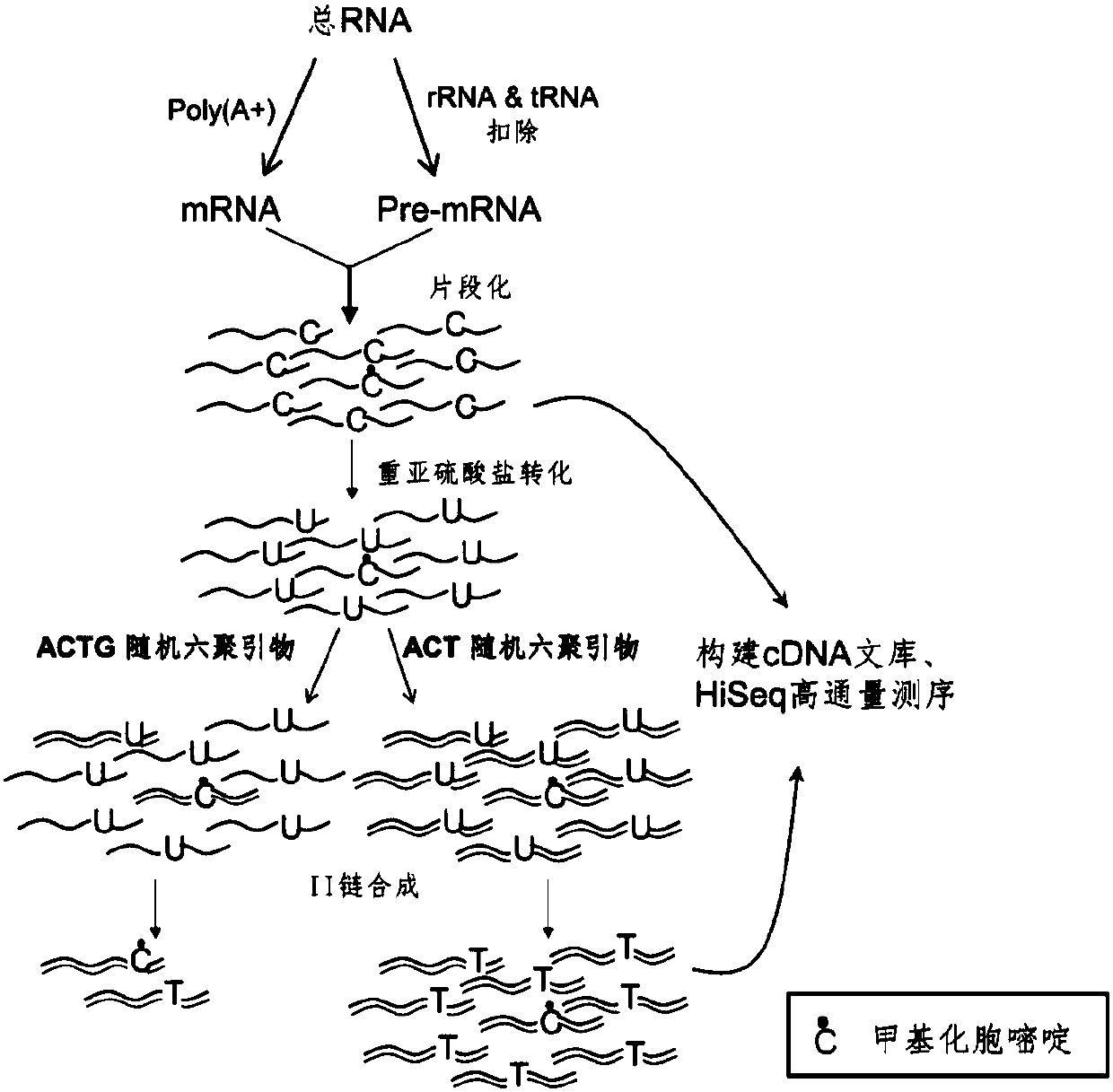
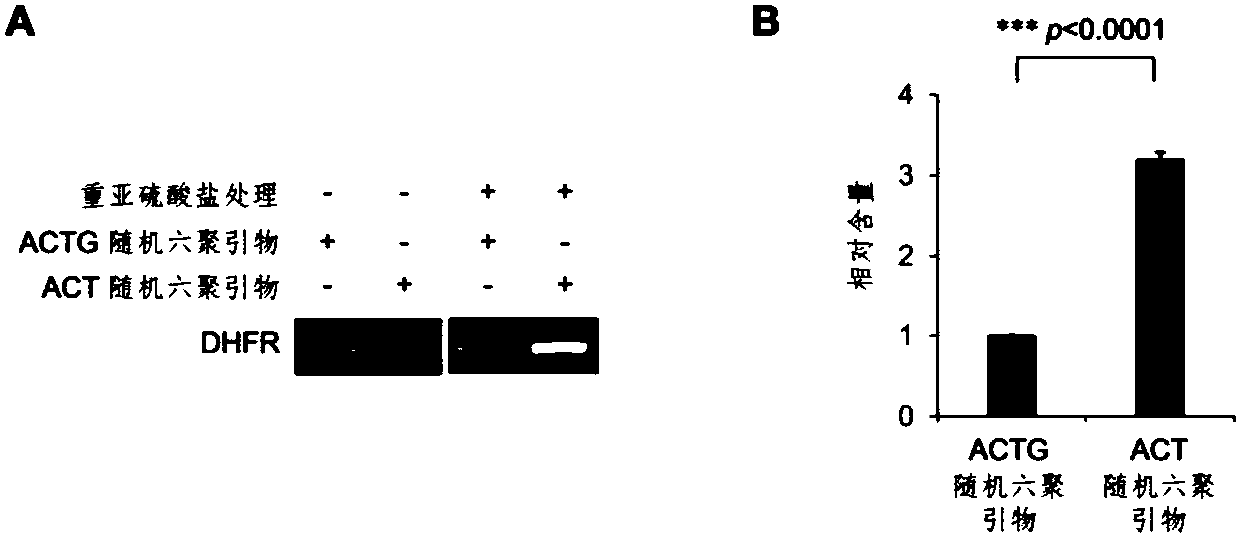
The invention relates to a library construction method, a sequencing method and a methylation detection method for sequencing 5-methylcytosine (5mC) modified bisulfite on RNA molecule. By designing and using triplet base random hexamers which contain ACT only, RNA fragment reverse transcription, PCR amplification efficiency and 5mC site detection efficiency after bisulfite treatment can be significantly improved, which is conducive to high-throughput sequencing of the 5mC site.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
Method and device for simultaneously detecting methylation level, genome variation and insert fragment
ActiveCN111755072AEasy to distinguishImprove the comparison rateProteomicsGenomicsGenes mutationBisulfite sequencing
The invention provides a method for simultaneously detecting a methylation level, genome variation and an insert fragment by utilizing a methylation non-bisulfite sequencing technology. The genome variation comprises gene mutation, copy number variation and structural variation. The invention also provides a device and equipment for implementing the method, and a corresponding computer readable medium. According to the invention, for methylated non-bisulfite sequencing data, one-step detection and analysis from offline data to multiple dimensions of methylation level, genome variation and insert fragments are realized, the method is suitable for whole genome and targeted capture data types of methylated non-bisulfite, and analysis of a single cancer sample and paired samples (cancer samples containing control samples) can be carried out.
Owner:深圳吉因加医学检验实验室
Kit for morbidity-related tumor suppressor gene epigenetic mutation detection of gastrointestinal neoplasms and application thereof
The invention relates to a kit for morbidity-related tumor suppressor gene epigenetic mutation detection of gastrointestinal neoplasms and application thereof. The kit mainly comprises 25 pairs of gastrointestinal neoplasm morbidity-related tumor suppressor gene promotor region CpG-island amplification and sequencing primers of 9 kinds and a polymerase chain reaction (PCR) amplification reagent. Through a PCR amplification product obtained through the kit, a gene CpG-island methylation state can be analyzed through the methods of methylation specific PCR (MSP), combined bisulphite-restrictionanalysis (COBRA) or bisulfite sequencing PCR (BSP) and the like. The kit has the efficient effect of detecting CpG-island methylation and is used for the morbidity-related tumor suppressor gene epigenetic mutation detection of the gastrointestinal neoplasms.
Owner:NANJING UNIV
Machine learning implementation for multi-analyte assay development and testing
Systems and methods that analyze blood-based cancer diagnostic tests using multiple classes of molecules are described. The system uses machine learning (ML) to analyze multiple analytes, for example cell-free DNA, cell-free microRNA, and circulating proteins, from a biological sample. The system can use multiple assays, e.g., whole-genome sequencing, whole-genome bisulfite sequencing or EM-seq, small-RNA sequencing, and quantitative immunoassay. This can increase the sensitivity and specificity of diagnostics by exploiting independent information between signals. During operation, the system receives a biological sample, and separates a plurality of molecule classes from the sample. For a plurality of assays, the system identifies feature sets to input to a machine learning model. The system performs an assay on each molecule class and forms a feature vector from the measured values. The system inputs the feature vector into the machine learning model and obtains an output classification of whether the sample has a specified property.
Owner:FREENOME HLDG INC
Methylation quantitative detection method of PCDH8 gene
InactiveCN101967515AQuantitative determination of abnormal methylation levelsSimple and fast operationMicrobiological testing/measurementReference genesBisulfite sequencing
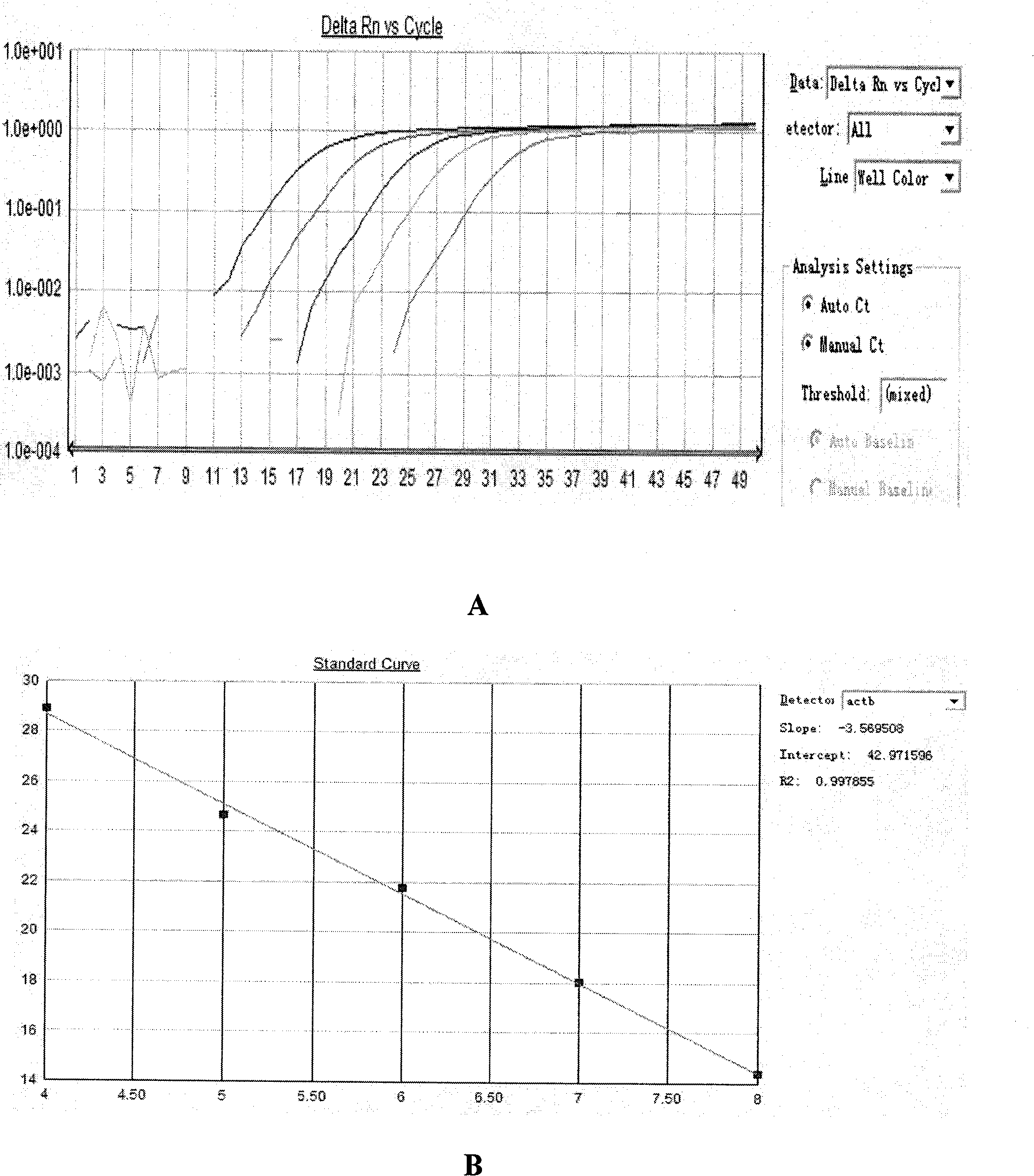
The invention belongs to the technical field of biomedicine. Based on the fact that the abnormal methylation of genes are involved in the generation and development of tumors, the invention aims to overcome the defects of a methylation sensitive restriction enzyme method, a bisulfite sequencing method and a methylation specificity PCR (Polymerase Chain Reaction) method, and provide a methylation quantitative detection method of the PCDH8 (Protocadherin-8) gene, with simple and convenient operation and high sensitivity. The detection method comprises the steps of: taking pancreatic cancer tissues, extracting DNA to prepare a standard curve sample of ACTB (Actin, Beta), and preparing a standard curve sample of PCDH8; extracting the DNA of a tissue to be detected, carrying out chemical decoration, designing a methylation primer and a probe for a CPG island in a human PCDH8 gene promoter area, and designing a BSP primer and a probe for a CPG island in a human reference gene ACTB promoter area, carrying out real-time quantitative PCR on the DNA subjected to the sulfite treatment by using a Taqman-MGB real-time quantitative PCR method, and calculating out a quantitative value on the methylation degree of a PCDH8 gene. The method has the advantages of rapidness, correctness and high sensitivity.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Cancer screening test kit
The present invention relates to a cancer biomarker molecule, a screening test kit and a detection method thereof. According to the present invention, analysis of methylating areas of specimen target genes PAX1, ZNF582, SOX1 and NKX6-1 are adopted, plural of segments of target gene-specific oligonucleotide primers or probes are designed, and the oligonucleotide probes are adopted to detect presence or absence of methylation of the target genes so as to further determine possibility of cancer occurrence; and the methylation state detection method comprises methylation-specific PCR (MSP), quantitative methylation-specific PCR (QMSP), bisulfite sequencing (BS), microarrays, mass spectrometer, denaturing high-performance liquid chromatography (DHPLC), pyrosequencing or Enzyme-linked immunosorbent assay (ELISA) and the like.
Owner:湖南宏雅基因技术有限公司
LRRC55 gene methylation quantitative detection method
InactiveCN101967514ASimple and fast operationIncreased sensitivityMicrobiological testing/measurementReference genesBisulfite sequencing
The invention belongs to the technical field of biomedicine. The abnormal methylation of genes participates in the occurrence and the development of tumors. The invention aims at overcoming the defects of the methylation sensitivity restriction enzyme method, the bisulfite sequencing method and the methylation specificity PCR (Polymerase Chain Reaction) method and providing an LRRC55 gene methylation quantitative detection method with easy and simple operation and high sensitivity. The method comprises the following steps of: taking a pancreatic cancer tissue, extracting DNA, preparing a standard curve sample of ACTB (Actin, Beta), and preparing a standard curve sample of LRRC55 by using a complete methylation template; extracting the DNA of a tissue to be tested, performing chemical modification, designing a methylation primer and probe aiming at the CpG island of a promoter region of the human LRRC55 gene, designing a BSP primer and probe aiming at the CPG island of a promoter region of human reference gene ACTB, performing real-time quantitative PCR on the DNA treated by sulfite by using the Tagman-MGB real-time quantitative PCR method, and calculating the quantitative value of the methylation degree of the LRRC55 gene. The method is rapid and accurate and is high in sensitivity.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Unicell simplified representative bisulfite sequencing method and kit
PendingCN105506109AEasy to coverNucleotide librariesMicrobiological testing/measurementEnzyme digestionResearch Object
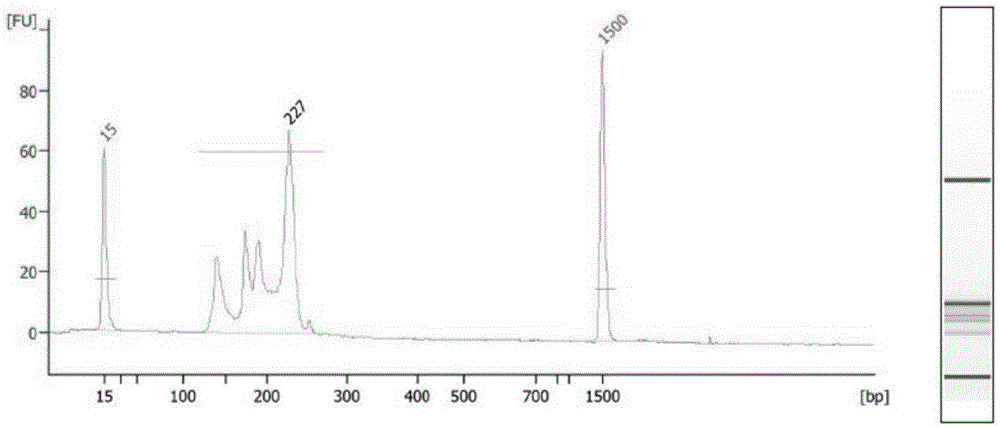
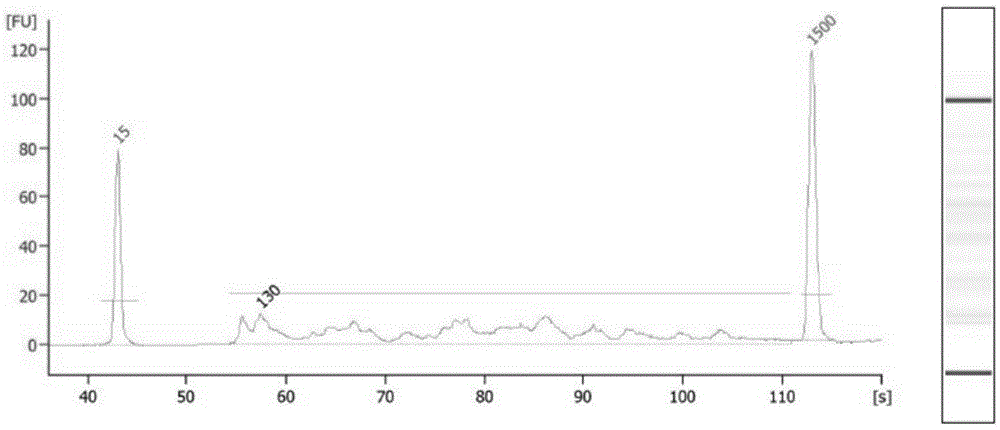
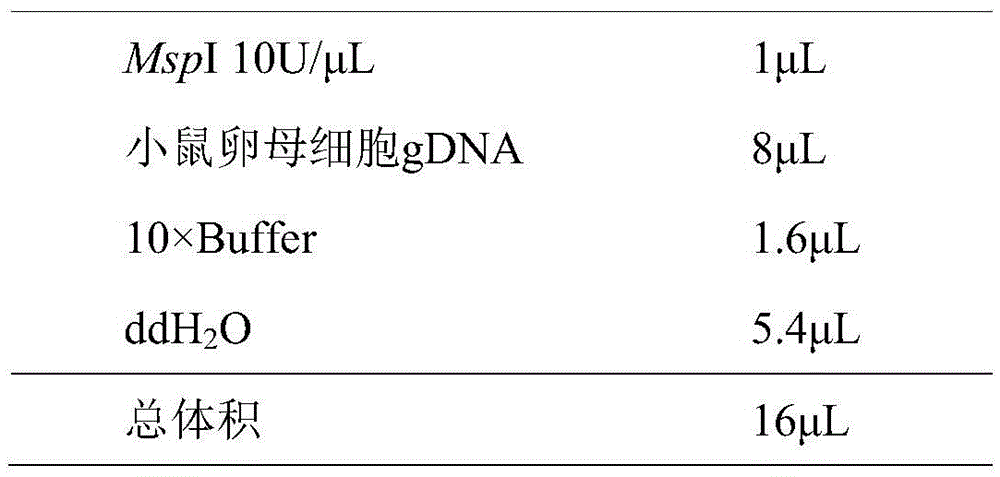
The invention relates to a unicell simplified representative bisulfite sequencing library construction method which comprises the steps of unicell pyrolysis, MspI enzyme digestion, tail end repair, 3'- terminal A addition, methylated linker addition, bisulfite treatment and PCR (polymerase chain reaction) amplification. The invention also relates to a unicell simplified representative bisulfite sequencing library construction kit, and a detection method and detection kit thereof, belonging to the field of gene sequencing. By using the unicells as the research object, the unicell simplified representative bisulfite sequencing library construction method for sequencing can be used for researching cell heterogeneity on the unicell level.
Owner:ANNOROAD GENE TECH BEIJING +2
Cancer screening method
InactiveUS7820386B2Accurate screening resultEasy diagnosisMicrobiological testing/measurementFermentationScreening cancerScreening method
A method for screening cancer comprises the following steps: (1) providing a test specimen; (2) detecting the methylation state of the CpG sequence in at least one target gene within the genomic DNA of the test specimen, wherein the target genes is consisted of SOX1, PAX1, LMX1A, NKX6-1, WT1 and ONECUT1; and (3) determining whether there is cancer or cancerous pathological change in the specimen based on the presence or absence of the methylation state in the target gene; wherein method for detecting methylation state is methylation-specific PCR (MSP), quantitative methylation-specific PCR (QMSP), bisulfite sequencing (BS), microarrays, mass spectrometer, denaturing high-performance liquid chromatography (DHPLC), and pyrosequencing.
Owner:NAT DEFENSE MEDICAL CENT
Method for detecting mycoplasma pneumonia (MP) of sheep
InactiveCN102363804AVerify dependenciesScreen sensitive and efficientMicrobiological testing/measurementMicroorganism based processesSusceptible individualBisulfite sequencing
The invention which belongs to the technical field of biological detection relates to a method for detecting MP of sheep and provides the methylation state of CpG islands in an MBL (mannan-binding lectin) gene promoter region related with the MP of sheep and an application thereof. The invention also provides a method for detecting the MP of sheep and concretely relates to the methylation detection of the MBL promoter region and the artificial infection verification. The methylation state detection of the MBL gene promoter region of sheep is carried out through selecting a bisulfite sequencing process, the methylation state and the frequency of each CpG locus in a given region can be accurately detected, three substantially differential CpG methylation loci, CpG3, CpG4 and CpG5, related with the MBL level are found, a case that the MBL level after toxic elimination is lower than the MBL level before the toxic elimination is promoted, and the MBL gene promoter region methylation may be a more substantial characteristic of the MBL level reduction of serum. The method allows the MP infection of sheep and susceptible individuals to be sensitively and accurately detected, and provides complete information and scientific bases for the sheep MBL gene promoter region methylation and the MP infection research of sheep.
Owner:赵宗胜
Methylated quantitative detection method of GSH2 (glutathione 2) gene
InactiveCN101818203ASimple and fast operationIncreased sensitivityMicrobiological testing/measurementReference genesBisulfite sequencing
The invention belongs to the technical field of biomedical sciences. Abnormal methylation of genes participates in the development of tumors. The invention aims at overcoming the defects of a methylation-sensitive restrictive endonuclease method, a bisulfite sequencing method and a methylation-specific PCR (Polymerase Chain Reaction) method and provides a methylated quantitative detection method of a GSH2 (glutathione 2) gene, which has simple and convenient operation and high senibilitiy. The method comprises the following steps of: taking pancreatic cancer tissues, extracting DNA and preparing an ACTB (Actin, Beta) standard curve sample, and preparing a GSH2 (Glutathione 2) standard curve sample by utilizing a full methylation template; extracting DNA of tissues to be detected, and carrying out chemical modification; designing methylation primers and probes aiming at CPG islands at a human GSH2 gene promoter region, designing BSP (Bisulfite) primers and probes aiming at CPG islands at a human reference gene ACTB promoter region, carrying out real-time quantitive PCR on DNA after bisulfite treatment by adopting a Taqman-MGB real-time quantitive PCR method, and calculating a quantitive value of the GSH2 gene methylation degree. The method is rapid, accurate and high in sensitivity.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Library construction method for whole genome methylation bisulfite sequencing
ActiveCN105349528AMicrobiological testing/measurementLibrary creationBiotin-streptavidin complexMagnetic bead
The invention provides a library construction method for whole genome methylation bisulfite sequencing. The method is suitable for a medium initial sample size (5-200 ng). The method comprises the steps of obtaining fragmented genome DNAs from 5-200 ng genome DNAs; conducting bisulfite treatment on the fragmented genome DNAs to obtain a single-stranded bisulfite treatment product; synthesizing a first chain by means of an oligo1 primer marked with biotin with the single-stranded bisulfite treatment product as the template; digesting single-stranded DNAs in a system after the first chain is synthesized with exonuclease; obtaining the first chain marked with biotin by means of streptavidin magnetic beads; synthesizing a second chain by means of an oligo2 primer with the first chain marked with biotin as the template; conducting PCR amplification with the second chain as the template, so that a DNA library for next-generation sequencing is constructed.
Owner:ZHEJIANG ANNOROAD BIO TECH CO LTD +2
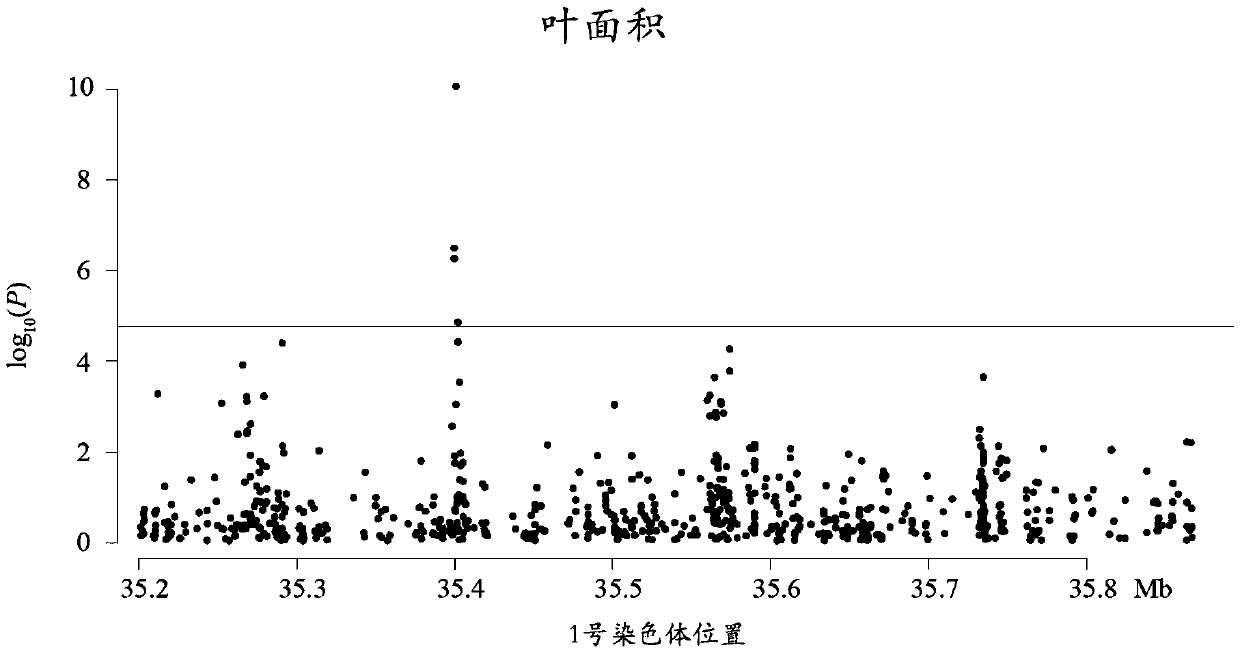
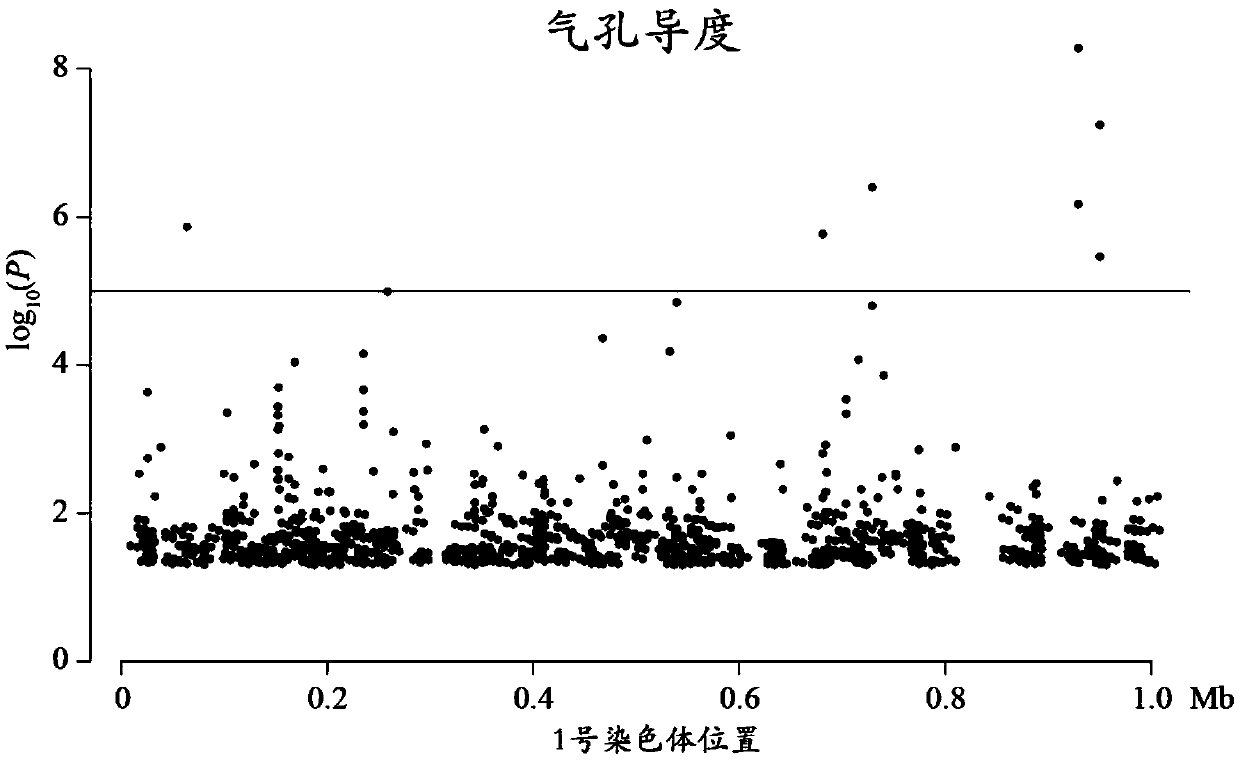
Method for detecting additive and dominant genetic effects of DNA methylation sites on quantitative traits and application technology thereof
InactiveCN109554502ANucleotide librariesMicrobiological testing/measurementPopulation sampleQuantitative trait locus
Owner:BEIJING FORESTRY UNIVERSITY
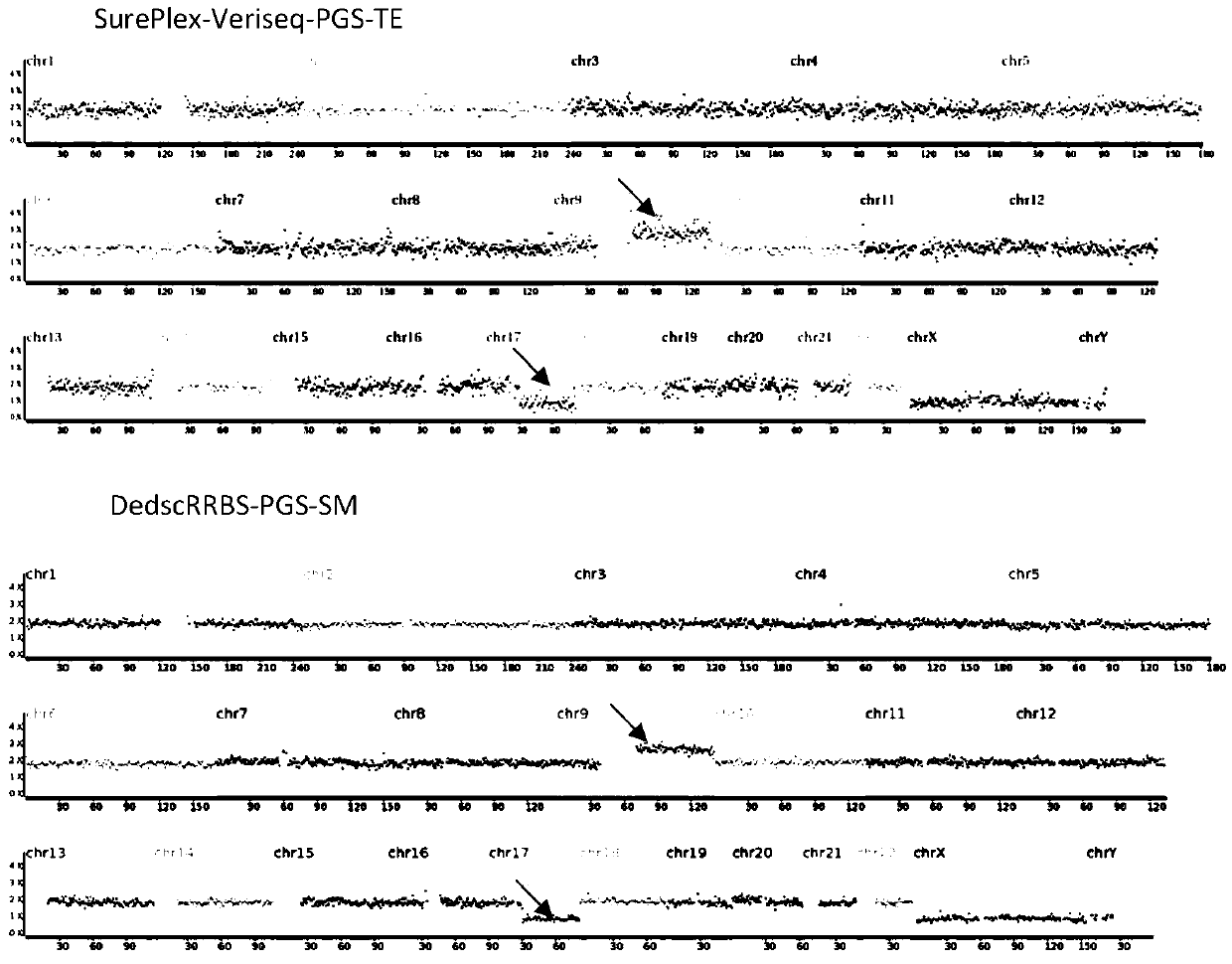
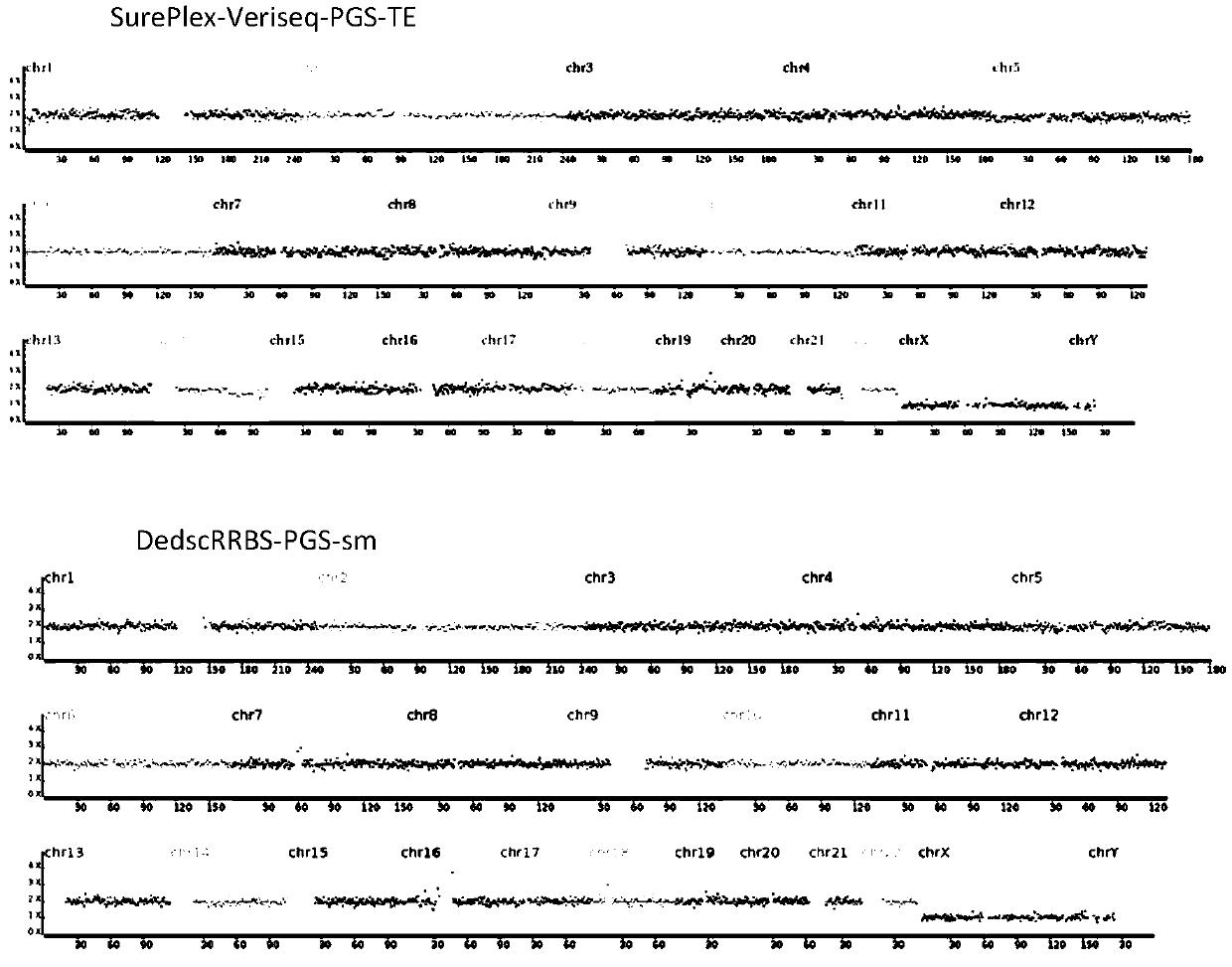
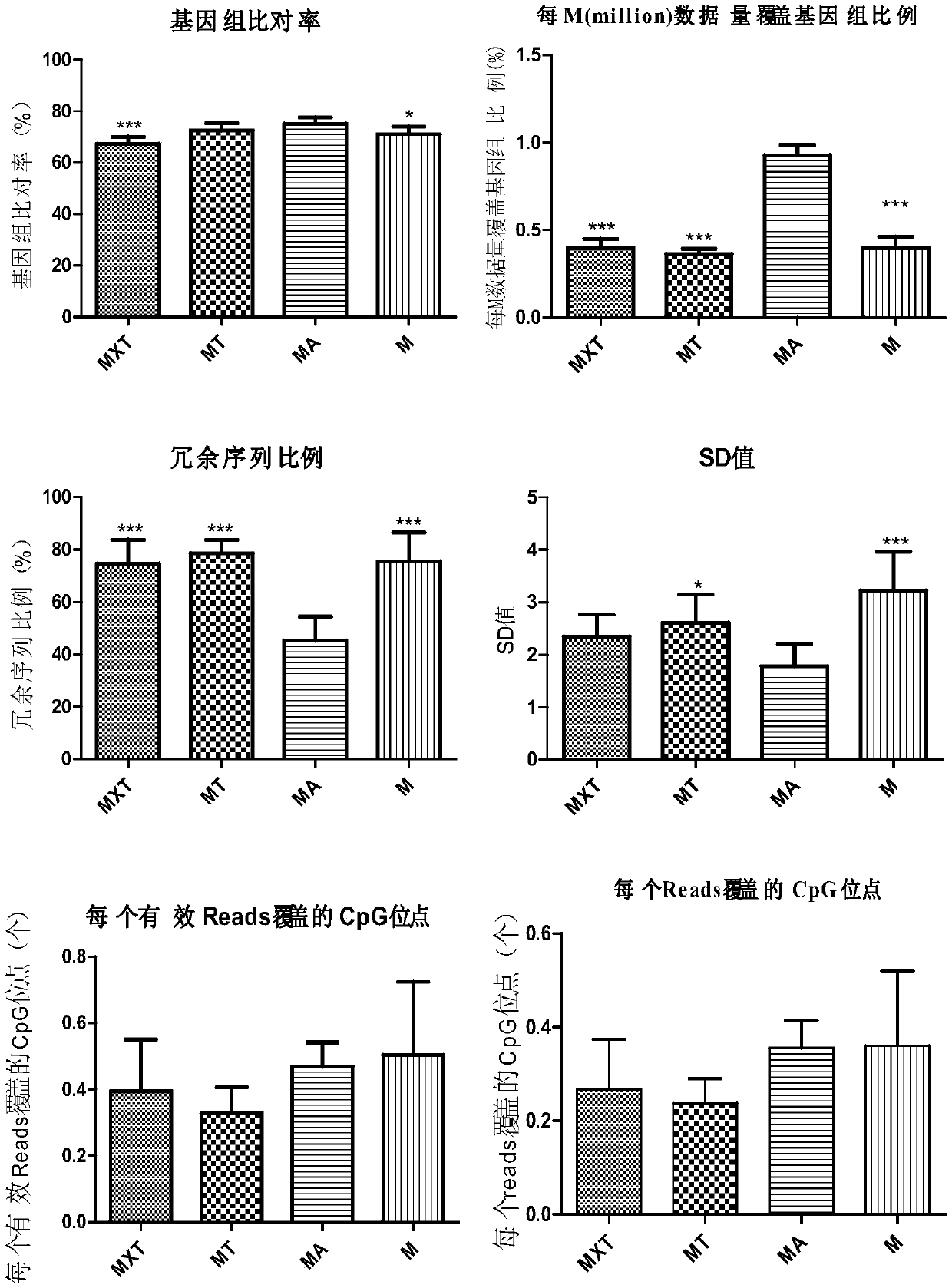
Method for performing DedscRRBS-PGS analysis on embryo culture solution
The invention discloses a method for performing DedscRRBS-PGS analysis on an embryo culture solution. The invention provides a method for performing simplifying representative bisulfite sequencing andchromosomal aneuploidy status analysis on the embryo culture solution, and the method comprises the steps of 1) constructing a DNA library by using a blastocyst culture solution as a sample, and in the process of constructing the DNA library, using two restriction enzymes to enrich CpG sites by digesting genomic DNA of the blastocyst culture solution; 2) conducting CT conversion on the DNA library with bisulfite to obtain a conversion product; 3) using the conversion product for preparing a methylation sequencing library, following by high-throughput sequencing, and analyzing DNA methylationand a chromosomal aneuploidy status based on a sequencing result. The blastocyst culture solution is used as a raw material, a double enzyme digestion simplifying representative bisulfite sequencing technology is adopted, and the development potential of an embryo is comprehensively evaluated from multiple angles.
Owner:北京中科遗传与生殖医学研究院有限责任公司 +1
Quantitative detection method of HPP1 gene methylation
InactiveCN101665835AQuantitative determination of abnormal methylation levelsSimple and fast operationMicrobiological testing/measurementFluorescence/phosphorescenceReference genesBisulfite sequencing
The invention belongs to the technical field of biomedicine. The abnormal methylation of the gene participates in the occurrence and the development of tumors, therefore, the invention aims at overcoming the defects of a methylation sensitive restriction enzyme method, a bisulfite sequencing method and a methylation specificity PCR method, and providing a quantitative detection method of HPP1 genemethylation with convenient operation and high sensitivity. The method comprises the steps of: taking pancreatic cancer tissue; extracting NDA to prepare a standard curve sample of ACTB; using a fullmethylated template to prepare a standard curve sample of HPP1; extracting the DNA of tissue to be tested, and chemically modifying; designing methylated primer and probe by aiming at the human HPP1gene promoter area CPG island; designing BSP primer and probe by aiming at the human reference gene ACTB promoter area CPG island; conducting real-time and quantitative polymerase chain reaction (PCR)to the DNA of processed sulfite by means of Taqman-MGB real-time quantitative PCR method; and counting the quantitative value of the HPP1 gene methylation degree. The method is fast and accurate, andhas high sensitivity.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Kit for morbidity-related tumor suppressor gene epigenetic mutation detection of gastrointestinal neoplasms and application thereof
The invention relates to a kit for morbidity-related tumor suppressor gene epigenetic mutation detection of gastrointestinal neoplasms and application thereof. The kit mainly comprises 25 pairs of gastrointestinal neoplasm morbidity-related tumor suppressor gene promotor region CpG-island amplification and sequencing primers of 9 kinds and a polymerase chain reaction (PCR) amplification reagent. Through a PCR amplification product obtained through the kit, a gene CpG-island methylation state can be analyzed through the methods of methylation specific PCR (MSP), combined bisulphite-restrictionanalysis (COBRA) or bisulfite sequencing PCR (BSP) and the like. The kit has the efficient effect of detecting CpG-island methylation and is used for the morbidity-related tumor suppressor gene epigenetic mutation detection of the gastrointestinal neoplasms.
Owner:NANJING UNIV
Methylation gene for liver cancer screening
ActiveCN106811523ASensitive detectionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideCervical cancer screening
The invention discloses a methylation gene for liver cancer screening. The gene is STAP1, and the nucleotide sequence of the gene is as shown in SEQ ID No: 1 and at least contains a methylation locus. According to the methylation gene for liver cancer screening, methylation sequencing can be performed by the aid of methods such as a methylation specific polymerase chain reaction method, a bisulfite sequencing method, a micro-array method, a mass-spectrographic analysis method, a denaturing high-performance liquid chromatography method and a pyrosequencing method, and the methylation gene is high in sensitivity, reliable in result and high in practicability and can sensitively detect early-phase liver cancer and earlier-phase liver cancer.
Owner:BEIJING YOUAN HOSPITAL CAPITAL MEDICAL UNIV
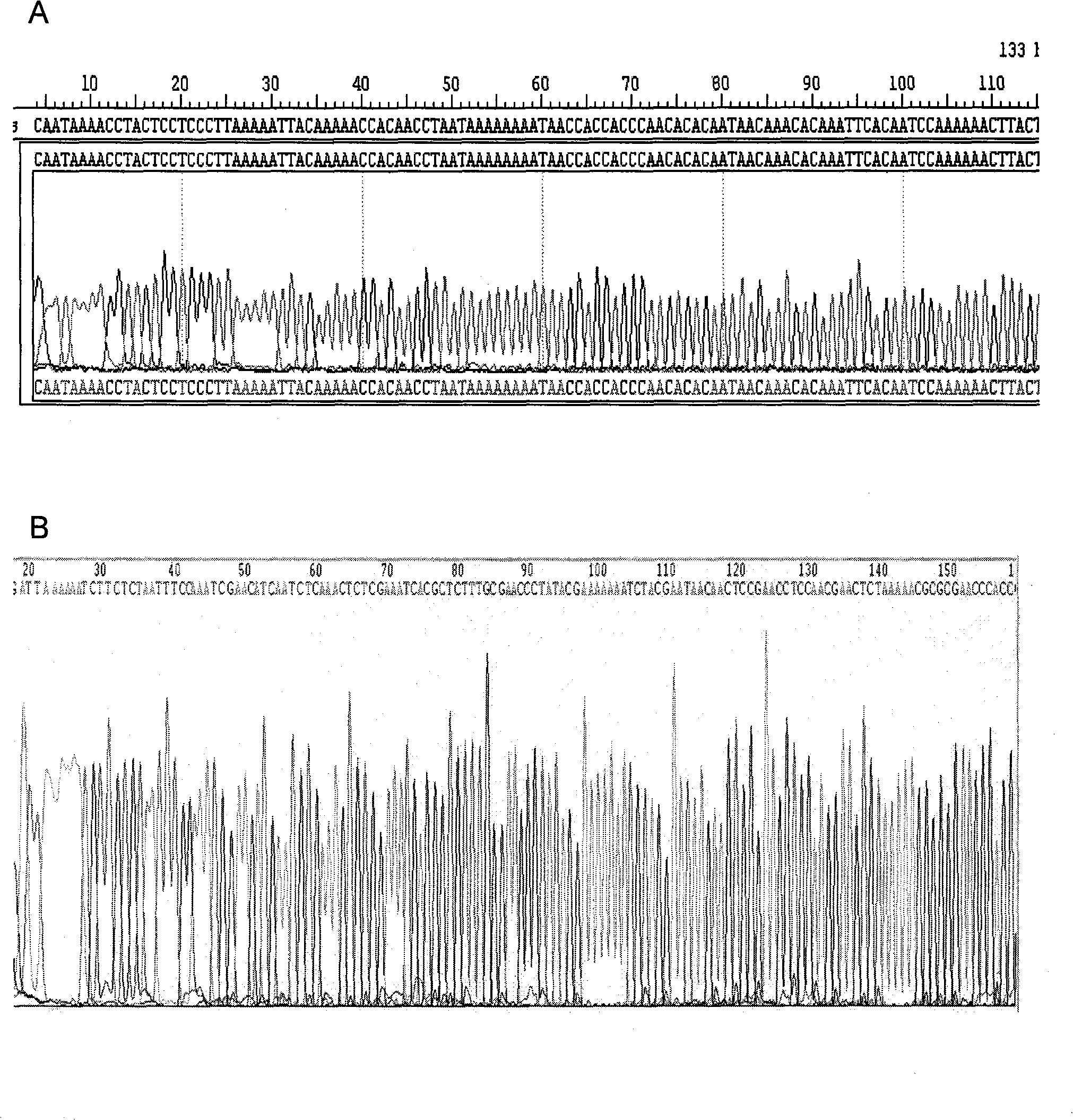
FOXP3 (forkhead/winged helix transcription factor 3) gene methylation detection method based on bisulfite sequencing
InactiveCN107236805AImprove accuracyGood repeatabilityMicrobiological testing/measurementLiquid mediumKanamycin
The invention provides an FOXP3 (forkhead / winged helix transcription factor 3) gene methylation detection method based on bisulfite sequencing. The method comprises the following steps: 4 mL of human venous blood is collected by a coagulation-promoting tube, solidified at room temperature for 30 min and centrifuged at 1,000 g*15 min; genomic DNA of a blood clot is extracted; integrity, concentration and purity of the genomic DNA are determined primarily; the genomic DNA is modified with a methylation kit; methylation PCR amplification is performed, a PCR product is identified with 2% agarose gel electrophoresis, and a gel imaging system performs observation and shooting; 18 mu L of the PCR product is purified and recovered by a DNA purifying and recovering kit, 2 mu L of the purified and recovered PCR product is transformed into Trans1-T1 competent cells through vector linking, blue-white clone plaque selection is performed, and positive clone is identified; a kanamycin monoclonal antibody LB liquid medium with the concentration of 50 mu g / mL is inoculated with thalli in positive clone plaques, the thalli are subjected to shake culture at the constant temperature of 37 DEG C and the rate of 200 rpm / min for 8-12 h, and samples turning to be turbid are taken for sequencing. Methylation degrees of 5 CpG sites of a specific sequence of a promoter region of an FOXP3 gene can be detected successfully with the method.
Owner:深圳市龙岗区疾病预防控制中心 +1
Methylation high-flux detection method
ActiveCN102796808BComprehensive and accurate analysisSimple design methodMicrobiological testing/measurementLibrary creationHigh fluxBisulfite sequencing
Owner:BGI TECH SOLUTIONS
Cancer screening method
ActiveCN101864480BAccurate screening effectMicrobiological testing/measurementMethylation specific polymeraseScreening method
The invention relates to a cancer screening method which comprises the following steps that: (1) providing a tested body; (2) testing the CpG sequence methylation state of at least one target gene in genome DNA of the tested body, and the target gene comprises PTPRR, ZNF582, PDE8B and DBC1; (3) judging whether the tested body has cancer or lesions before cancer according to that whether the methylation state of the at least one target gene exists; and the methylation state detection method is methylation-specific PCR, MSP, quantitative methylation-specific PCR, QMSP, bisulfite sequencing (BS), microarrays, massspectrometer, denaturing high-performance liquidchromatography (DHPLC) and the like.
Owner:NAT DEFENSE MEDICAL CENT
A method of cancer screening
ActiveCN101570779BIncreased sensitivityStrong specificityMicrobiological testing/measurementScreening cancerBisulfite sequencing
A cancer screening method comprises the following steps: (1) a tested body is provided; (2) the methylation state of a CpG sequence of at least one target gene of genome DNA of the tested body is detected, wherein the target gene comprises SOX1, PAX1, LMX1A, NKX6-1, WT1 and ONECUT1; (3), whether the tested body has a cancer or a precancerous lesion is judged according to the methylation state of the target gene. A methylation state detection method adopts MSP (methylation-specific PCR), QMSP (quantitative methylation-specific PCR), BS (bisulfite sequencing), microarrays, mass spectrometer, DHPLC (denaturing high-performance liquid chromatography) and the like.
Owner:赖鸿政
Simplified method and kit for constructing library for apparent bisulfite sequencing
Owner:ANNOROAD GENE TECH BEIJING +3
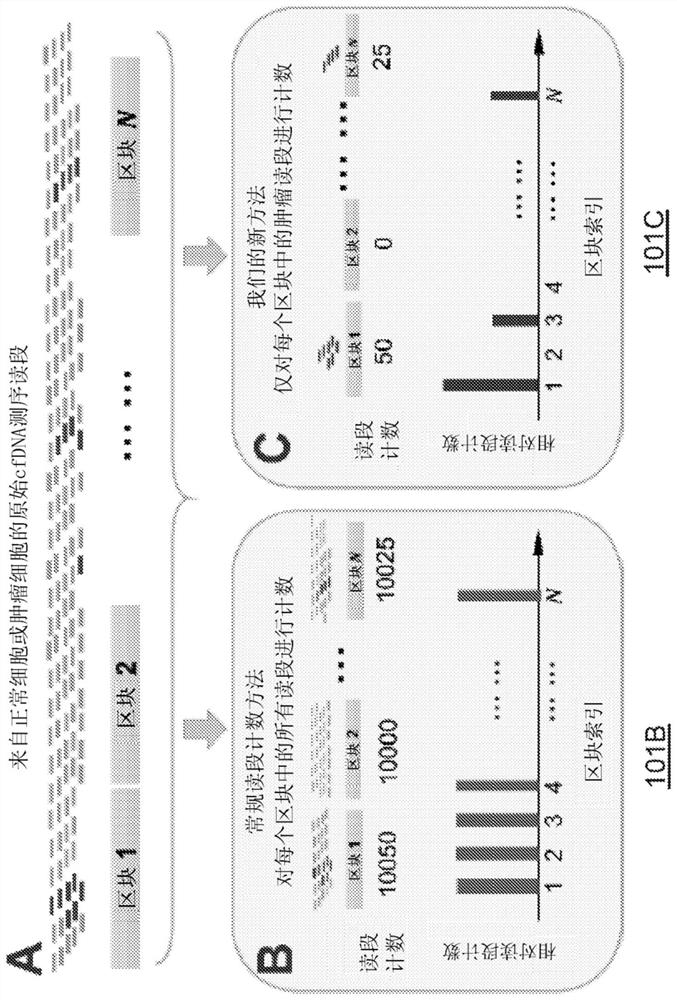
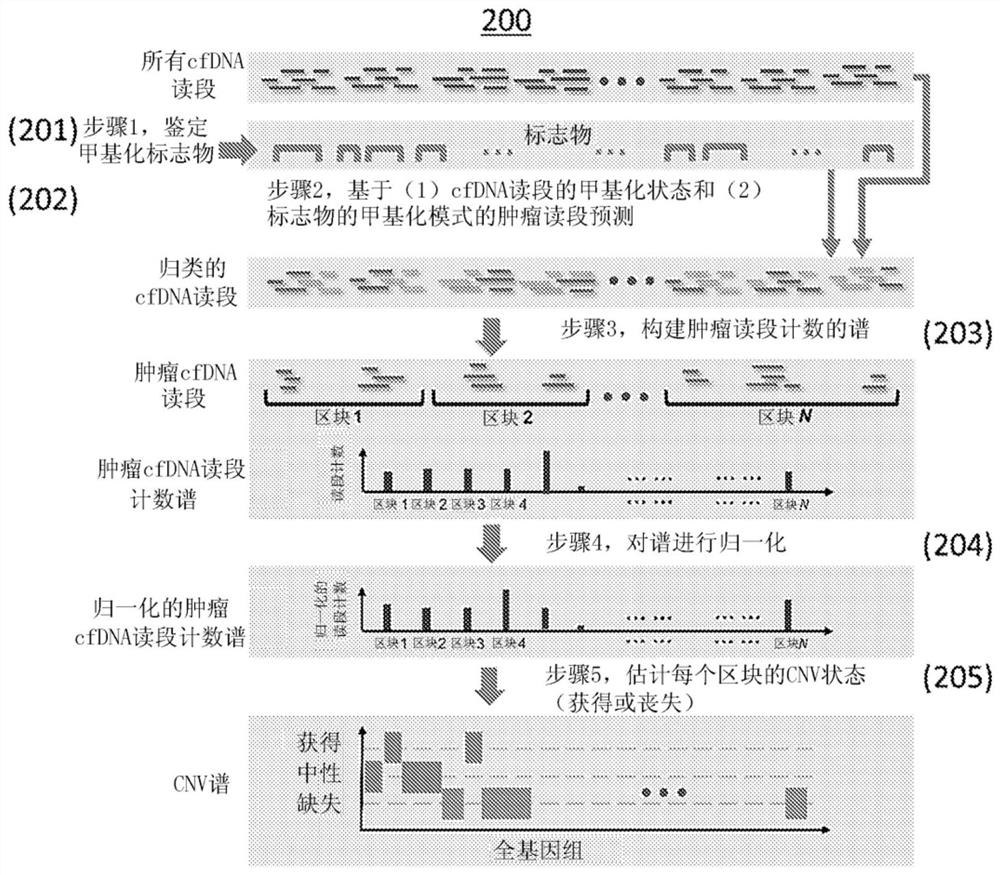
Sensitively detecting copy number variations (CNVs) from circulating cell-free nucleic acid
The present disclosure provides methods and systems for detecting or inferring levels of Copy Number Variants (CNVs) in cell-free nucleic acid samples to detect or assess cancer and prenatal diseases. Cell-free nucleic acid methylation sequencing data may be utilized to distinguish tumor-derived or fetal-derived sequencing reads from normal cfDNA sequencing reads. Each cell-free nucleic acid sequencing read (e.g., containing tumor or fetal methylation markers) may be classified as corresponding to a tumor / fetal-derived or a normal-plasma cell-free nucleic acid, based on the methylation cfDNA sequencing data (e.g., obtained using Bisulfite sequencing or bisulfite-free sequencing methods) and tumor / fetal methylation markers. Next, a profile of the tumor / fetal-derived sequencing read counts may be constructed and then normalized. The CNV status (e.g., gain or loss) of each genomic region may be inferred, and a diagnosis or prognosis can be made based on a subject's inferred CNV profile.
Owner:RGT UNIV OF CALIFORNIA +1
Library construction method and application of rna 5mC bisulfite sequencing
ActiveCN105132409BAvoid the disadvantage of high error rateImprove reverse transcription synthesis efficiencyMicrobiological testing/measurementLibrary creationBisulfite sequencingRandom hexamer
The invention relates to a library construction method, a sequencing method and a methylation detection method for sequencing 5-methylcytosine (5-methylcytosine, 5mC) modified bisulfite on RNA molecules. By designing and using a three-base random hexamer primer containing only ACT, the efficiency of reverse transcription and PCR amplification of RNA fragments treated with bisulfite and the detection efficiency of 5mC sites are significantly improved, which is beneficial to the detection of 5mC sites high-throughput sequencing.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
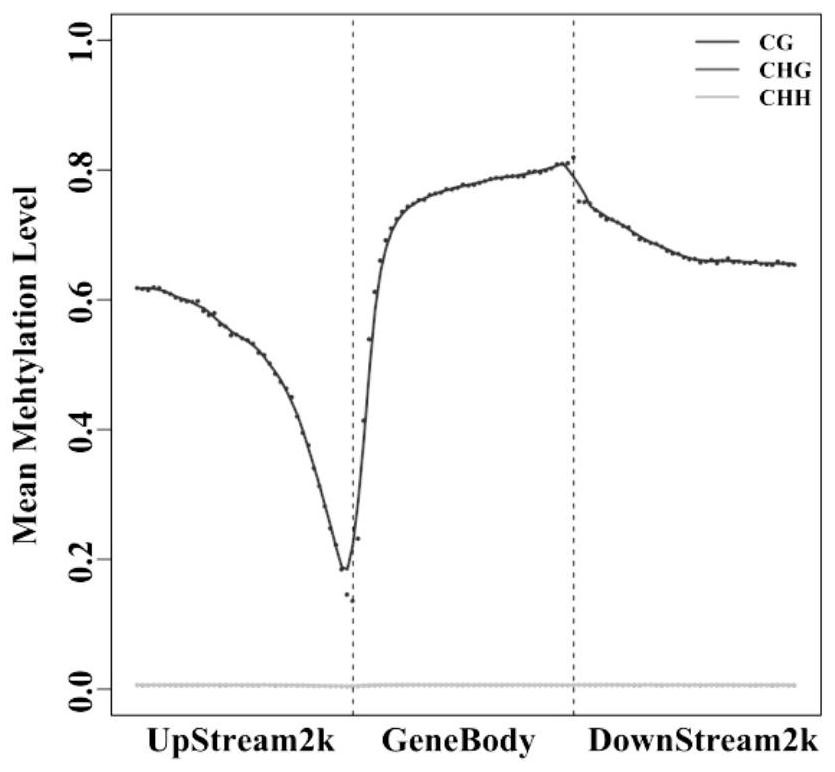
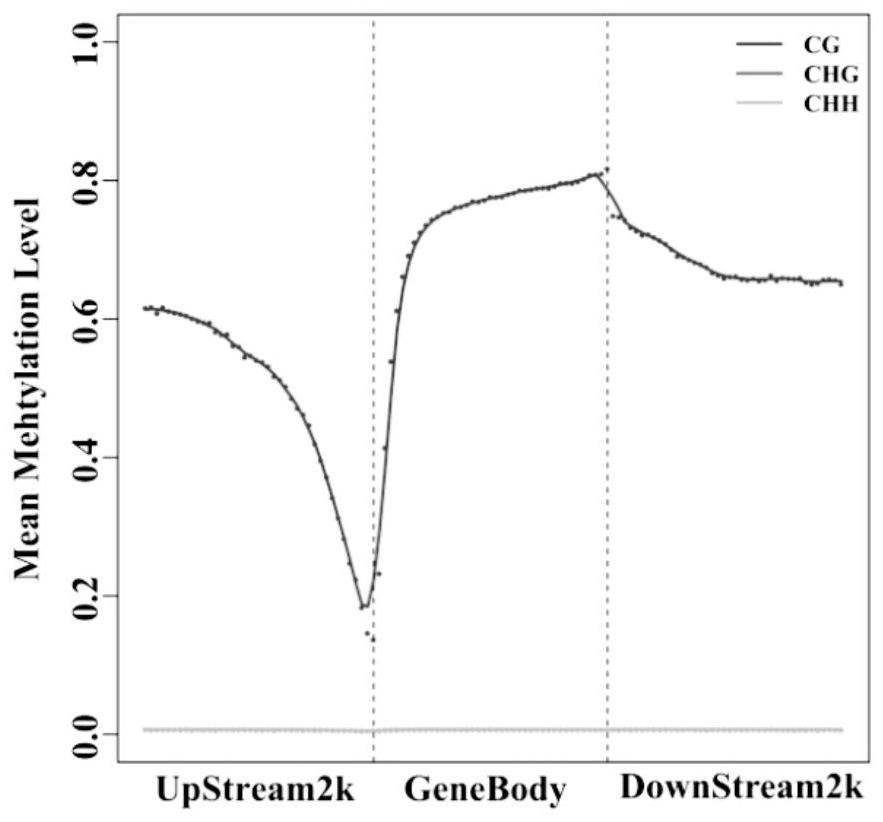
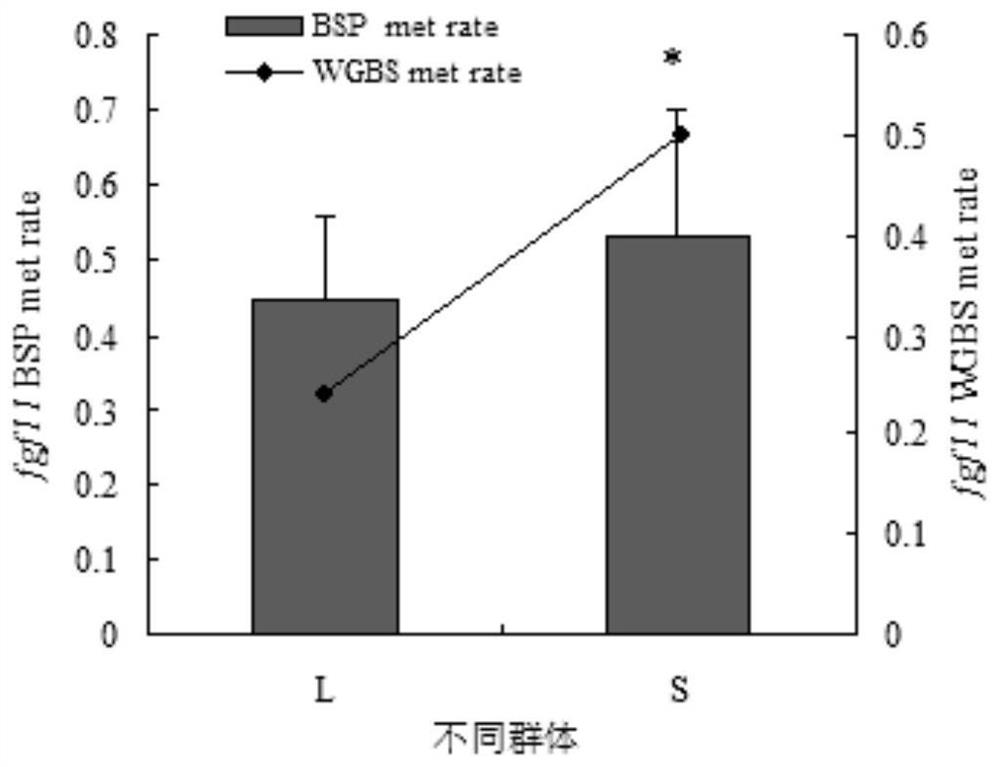
Molecular Markers of DNA Methylation in Large Yellow Croaker and Its Application in Breeding
ActiveCN113337617BImproved germplasmImprove breeding rateMicrobiological testing/measurementClimate change adaptationDNA methylationEpigenetics
Owner:NINGDE NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com