Cancer screening test kit
A technology for detecting sets and specimens, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, etc., and can solve problems such as obstacles to testing methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment one material and method
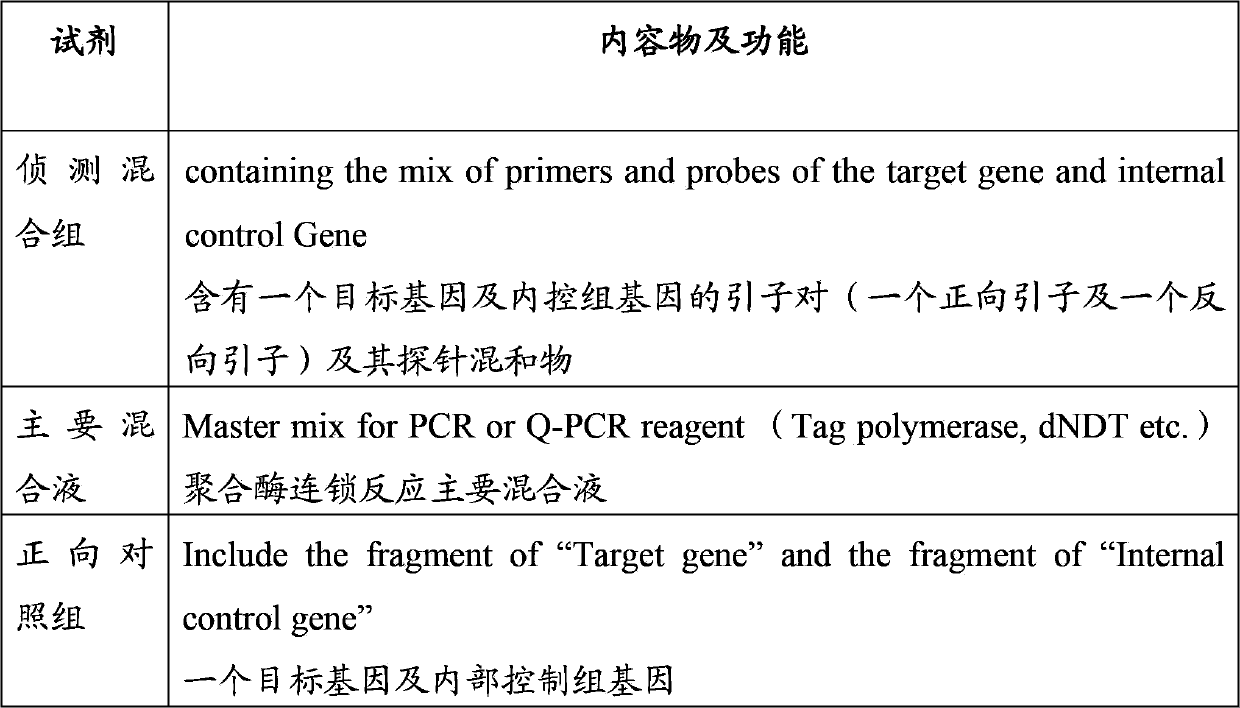
[0037] Samples, first extract their DNA, after the conversion of bisulfite, and reveal in Table 1 to Table 4 that it can be used to amplify the methylation of PAX1, ZNF582, SOX1, NKX6-1 and the internal control group Primer pairs and probe sets (such as Table 1 to Table 5) for the region are used for detection. The main components of the kit are shown in Table 6:
[0038] Table 1 Primer pairs and probe sets used to amplify the methylated region of the target gene PAX1
[0039] ID No.:
PAX1 primer pair and probe set
SEQ ID No: 1
5'attcgcgcgttttcggcgtga 3'
SEQ ID No: 2
5'gttaaattgattttcgtacgttgtag 3'
SEQ ID No: 3
5'tattttgggtttggggtcgc 3'
SEQ ID No: 4
5'ttattttgggtttggggtcgcg 3'
SEQ ID No: 5
5'gggcggtagcgcgtttcgtt3'
SEQ ID No: 6
5'tagcggcggcggtaggttttgga 3'
SEQ ID No: 7
5'gtagtgacgggaattaatgagt 3'
SEQ ID No: 8
5'aacatcccacgaccacgccg 3...
Embodiment 2
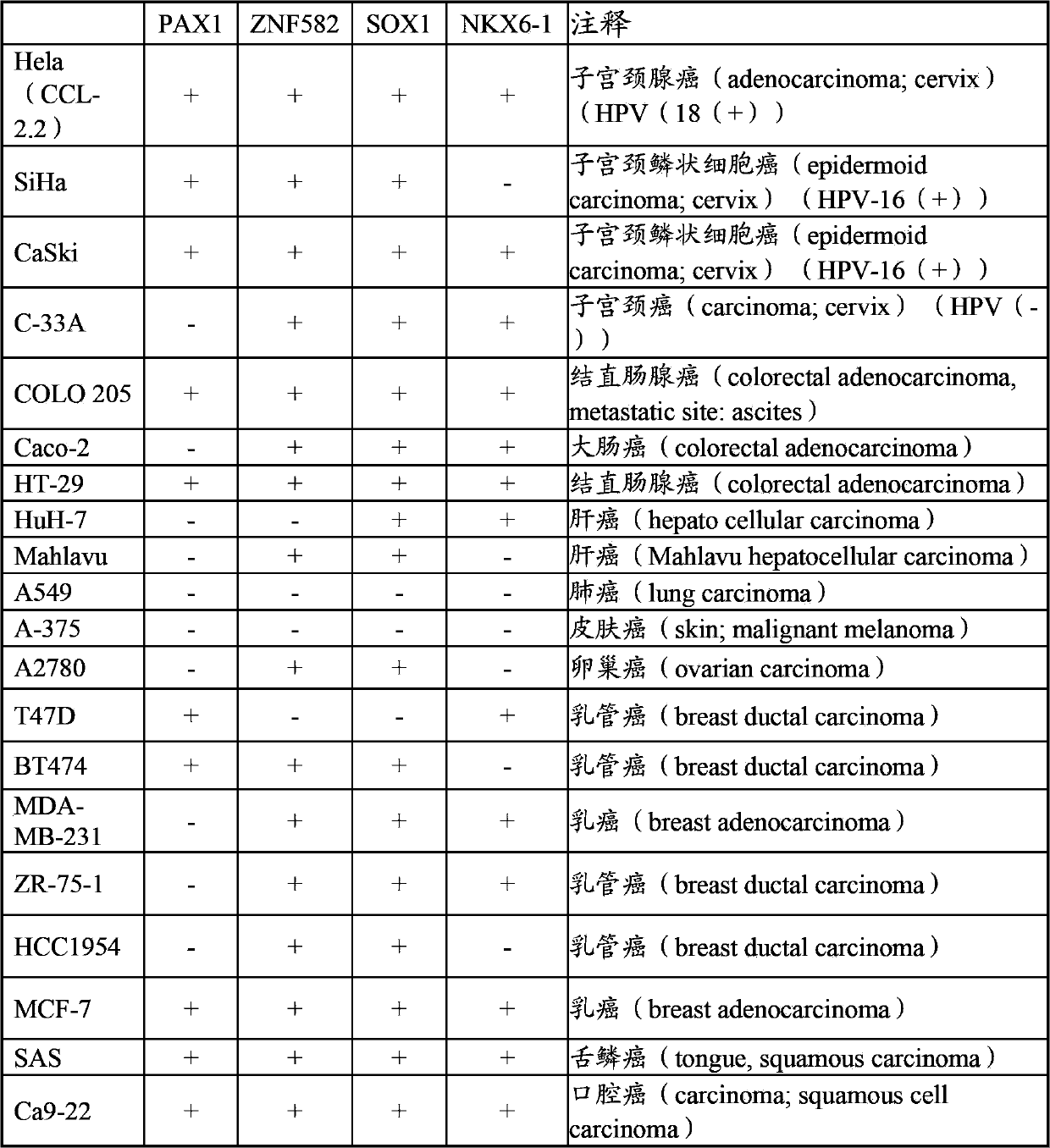
[0053] Example 2 Analysis of Methylation Degree of Target Genes in Different Cancer Cell Lines
[0054]This test uses the primer pairs and probe sets disclosed in Table 1 to Table 4 that can be used to amplify the methylated regions of PAX1, ZNF582, SOX1 and NKX6-1 to detect different cancer cell lines and analyze their methylation status The results are shown in Table 7. The results showed that in the cervical cancer-related cell line Hela, PAX1, ZNF582, SOX1 and NKX6-1 were all detected to be methylated; in the cervical cancer-related cell line SiHa, PAX1, ZNF582 and SOX1 were detected There is methylation; in the cell line CaSki related to cervical cancer, methylation was detected in PAX1, ZNF582, SOX1 and NKX6-1; in the cell line C-33A related to cervical cancer, ZNF582, SOX1 and NKX6 -1 Methylation detected. In the colorectal cancer-related cell line COLO 205, methylation was detected in PAX1, ZNF582, SOX1, and NKX6-1; in the colorectal cancer-related cell line Caco-2, ...
Embodiment 3
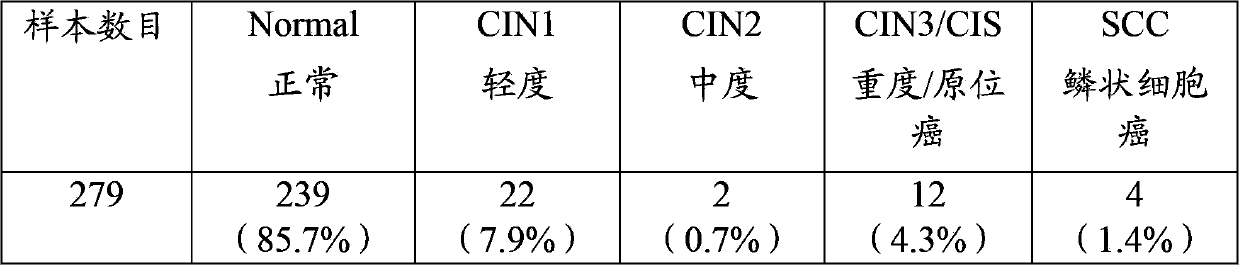
[0058] Example 3 Analysis of Methylation Degree of Target Genes in Cervical Cancer Samples
[0059] This test uses 279 normal and diseased Pap smear samples from Taiwan with known diagnoses. CIN2) 2 (0.7%), severe (CIN3) / carcinoma in situ (CIS) 12 (4.3%), squamous cell carcinoma 4 (1.4%), and their DNA was extracted, after bisulfite ( bisulfite), and use the primer pairs and probe sets disclosed in Table 1 to Table 4 that can be used to amplify PAX1, ZNF582, SOX1 and NKX6-1 methylated regions for detection. As shown in Table 9, compared with normal Pap smear samples, the Pap smear samples with PAX1, ZNF582, SOX1 and NKX6-1 as the target genes were 85.45 times more detected (95% confidence interval = 33.95~ 215.11), 289.17 times (95% confidence interval = 39.20-2133.14), 67.69 times (95% confidence interval = 20.55-223.01) and 2.56 times (95% confidence interval = 1.36-4.82) times the risk of developing severe cervical cancer. As shown in Table 10, when individual methylation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com