Library construction method for RNA 5mC bisulfite sequencing and application of library
A bisulfite and sequencing library technology, applied in the application field of 5mC high-throughput sequencing, can solve the problems affecting the exon retention level transcript assembly form, etc., to improve the efficiency of reverse transcription synthesis, comprehensive and complete detection , the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
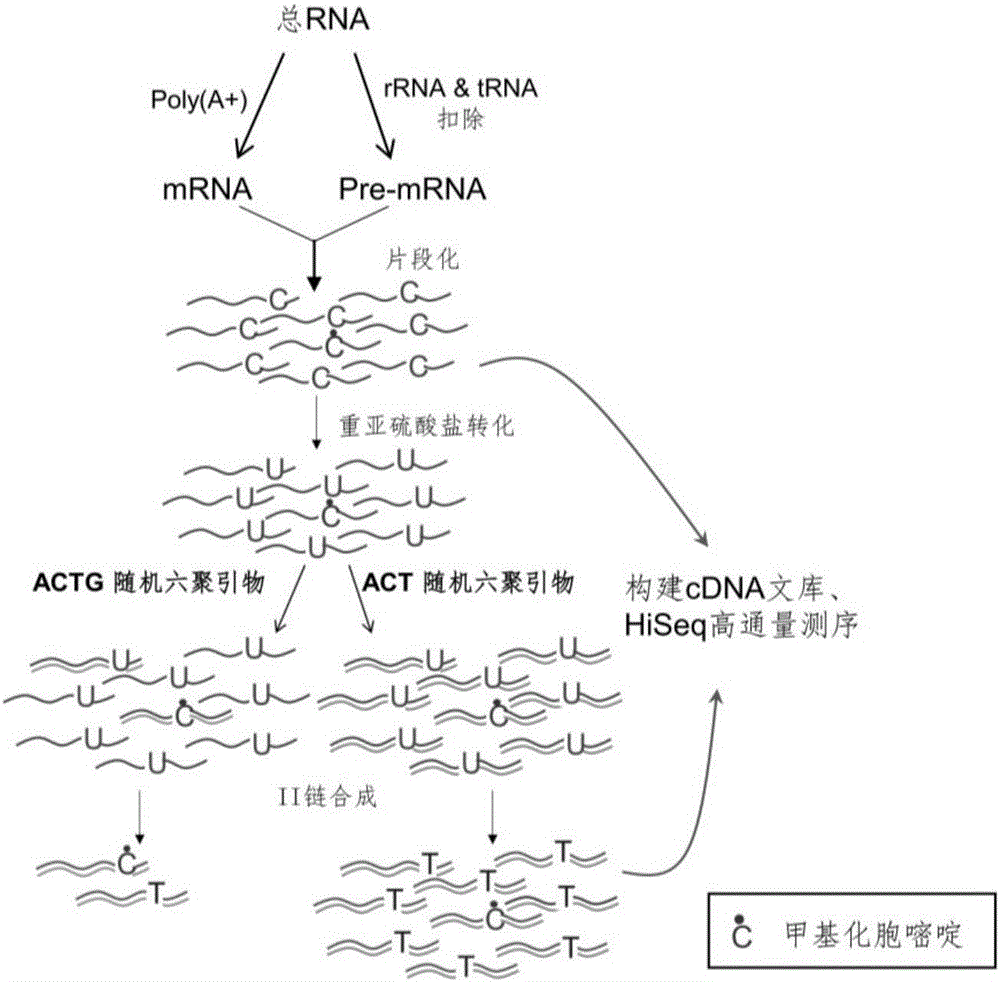
[0083] 1) RNA extraction and fragmentation
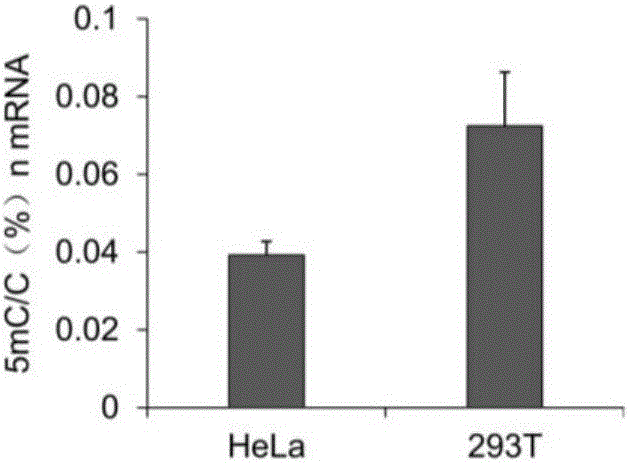
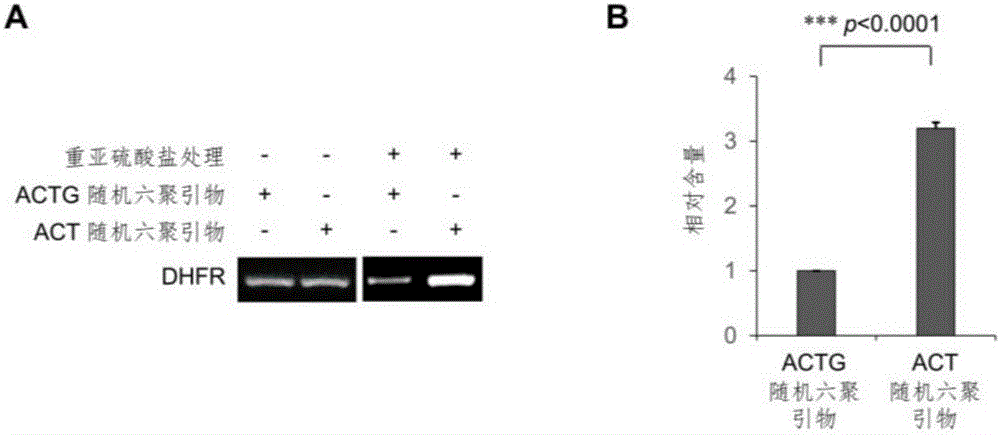
[0084] Using T7 in vitro transcription kit (NEB, E2040S) to transcribe the RNA sequence of DHFR from the plasmid pcDNA3-HA-mDHFR carrying the full length of the mouse DHFR gene, and use it as a standard for identifying the conversion efficiency of bisulfite. Total RNA was extracted from human HeLa cells by Trizol method. according to mRNAPurificationKit (Ambion, 61006) shows that mRNA is isolated and purified from total RNA. Mix 2 μg mRNA and 20 ng DHFR-RNA standard obtained by in vitro transcription to form an mRNA sample, and fragment the mRNA sample into 100 nt fragments according to the method instructions of the Ambion RNA Fragmentation Kit. The fragmentation condition was 2 μg mRNA / 9 μl for each reaction, 1 μl of fragmentation reagent was added, reacted at 90° C. for 1 minute, and then 1 μl of stop buffer was added. Glycogen, 3M sodium acetate (pH5.2) and pre-cooled pure ethanol were added to the reacted mRNA product, and ...
Embodiment 2
[0099] Download the human genome annotation file (version number 72) from the Ensembl database. The methylation sites obtained by sequencing analysis were annotated with Bedtools software based on the downloaded annotation files. The genes containing 5mC modification in the annotation results were classified based on the classification of genes in ensembl. The results showed that 87% of the genes containing 5mC methylation belonged to protein-coding genes ( Figure 4 A), the remaining modified genes are non-coding genes, including pseudogene, lincRNA, antisense, processedtranscript, etc. Among them, there are 202 5mC modified pseudogenes, 62 lincRNAs, 58 antisenses, and 39 processed transcripts ( Figure 4 B).
Embodiment 3
[0101] In order to examine the distribution of 5mC modifications in various regions of the transcript, the transcript was first divided into four regions according to the annotation information of the Ensembl database, 5' non-coding region (5'UTR), protein coding region (CDS), internal Intron (Intron) and 3' non-coding region (3'UTR). The positional information of the 5mC modification site on the mRNA in Example 1 was extracted, analyzed by Bedtools software and the annotation information downloaded from the Ensembl database, and detected that these methylation sites were in four regions of the mRNA (5'UTR, CDS, Intron and 3'UTR), the results showed that the distribution of these methylation sites in CDS was 35%, in introns was 28%, in 3' non-coding regions was 24%, and in 5' non-coding regions was 13% ( Figure 5 A). Calculate the distribution ratio of the C sites (coverage times greater than or equal to 1) on the mRNA measured in the sequencing results in the four regions ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| conversion efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com