FOXP3 (forkhead/winged helix transcription factor 3) gene methylation detection method based on bisulfite sequencing
A bisulfite and detection method technology, applied in the field of FOXP3 gene methylation detection, can solve the problems of low success rate and achieve the effect of improving accuracy and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The embodiments of the present invention will be described in detail below with reference to the accompanying drawings, but the present invention can be implemented in many different ways defined and covered by the claims.
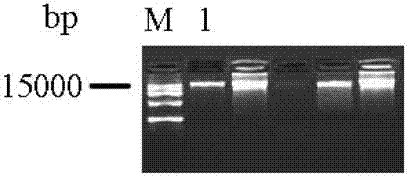
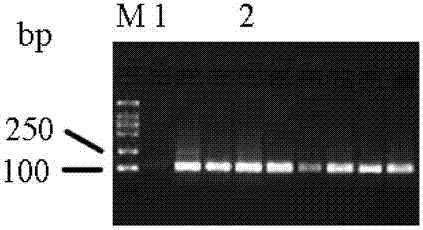
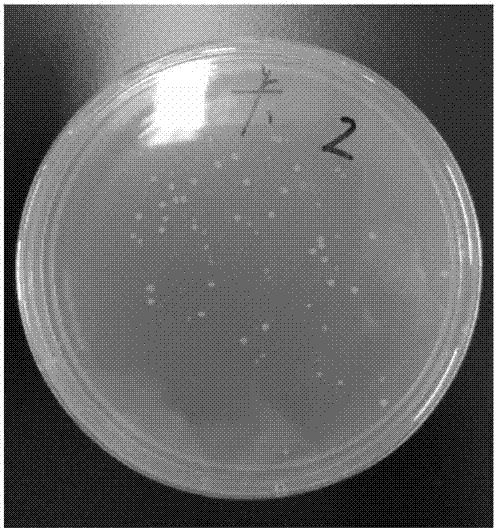
[0049] FOXP3 is a characteristic marker of Treg cells and plays an important role in maintaining immune tolerance. Recently, it has been found that the expression of FOXP3 gene and the function of the protein are all manifested as regulatory mechanisms other than DNA sequence variation, namely epigenetics, including DNA methylation, histone modification and post-translational modification. DNA methylation occurs most frequently with cytosine methylation. Cytosine methylation refers to the reaction in which a methyl group is added to the 5' position of cytosine to form 5-methylcytosine under the catalysis of DNA methyltransferase with S-adenosylmethionine as the donor. Part of the site is a CpG site formed by phosphate-linked cytosine and guanine. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com