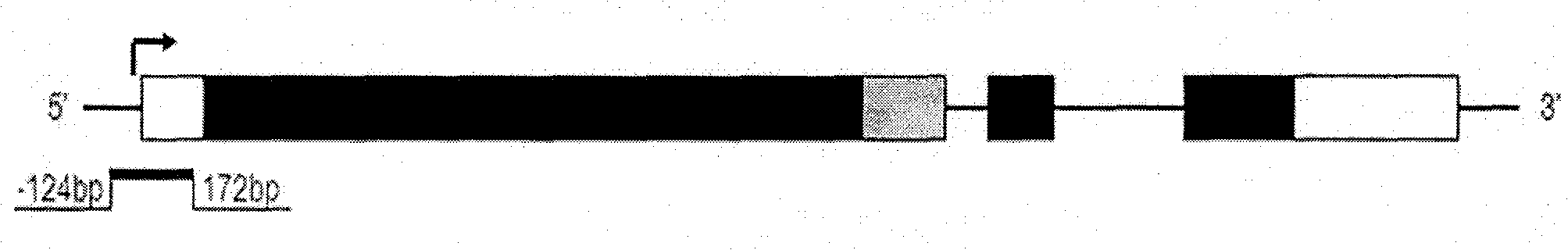
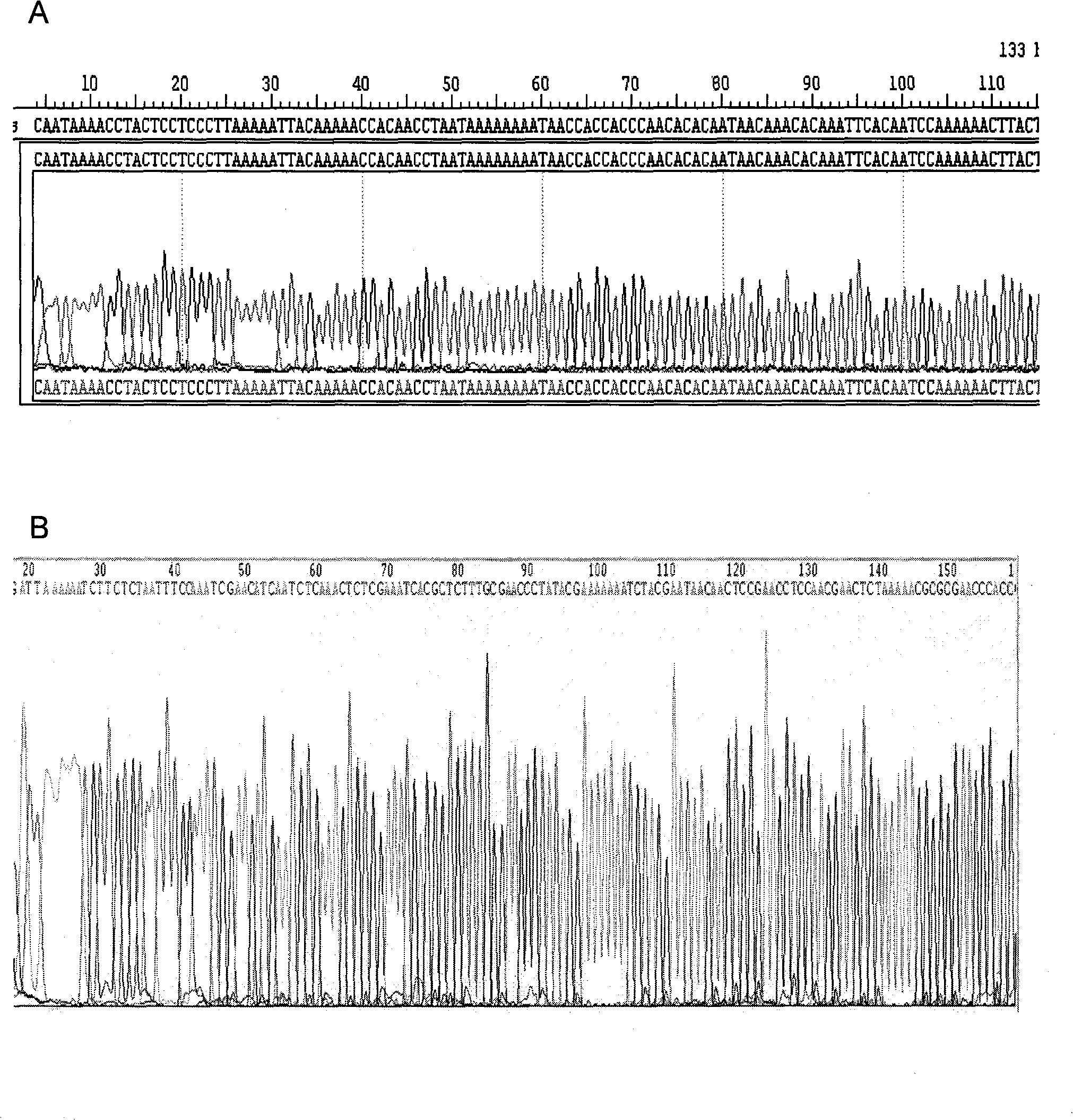
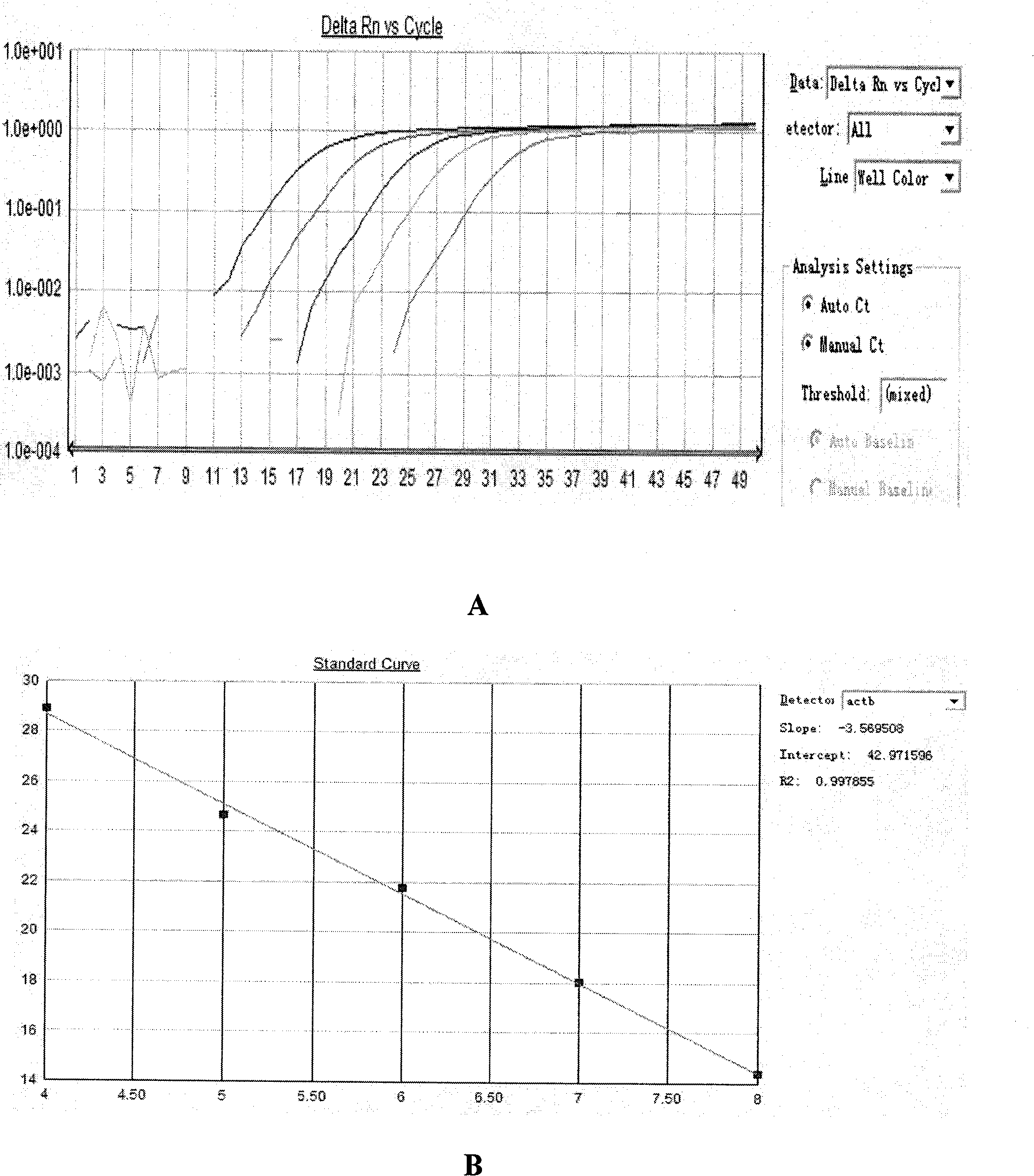
Methylation quantitative detection method of PCDH8 gene
A methylation quantification and detection method technology, applied to the field of PCDH8 gene methylation detection, can solve the problems of large sample size, complexity, CG neglect, etc., and achieve the effects of simple operation, high sensitivity and strong accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Quantitative detection of methylation degree of PCDH8 gene (GeneID: 5100) in clinical samples of patients
[0026] Sources of main reagents and instruments: QIAGEN EpiTect Bisufite KitTM kit, pMD18-TVector and EX-taq enzymes were purchased from Treasure Bioengineering (Dalian) Co., Ltd., ABI Taqman GeneExpression Master Mix was purchased from Applied Biosystems, Inc., and ABI 7500 Real Time PCR instrument Primers were synthesized by Shanghai Bioengineering Technology Service Co., Ltd. in Applied Biosystems, USA.
[0027] Steps:
[0028] 1. Preparation of standard products using gene amplification and bioengineering methods
[0029] 1. Genomic DNA Extraction
[0030] DNA was extracted by conventional phenol / chloroform method.
[0031] (1) Put 50mg of pancreatic tissue into a 5ml centrifuge tube, add 700ul of lysis buffer to make a homogenate.
[0032] (2) Add proteinase K to a final concentration of 100ug / ml, and keep at 55°C overnight until the solution bec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com