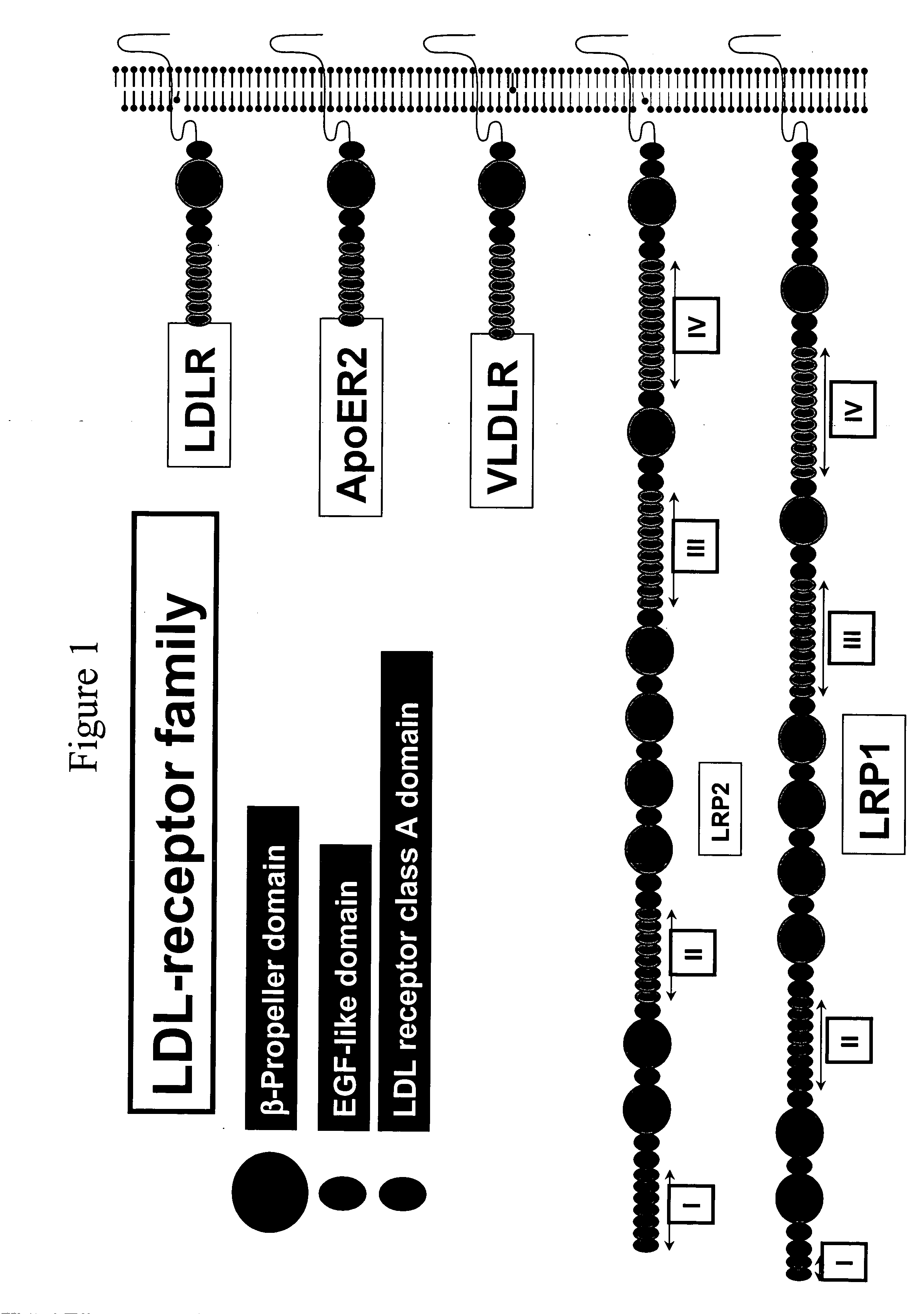
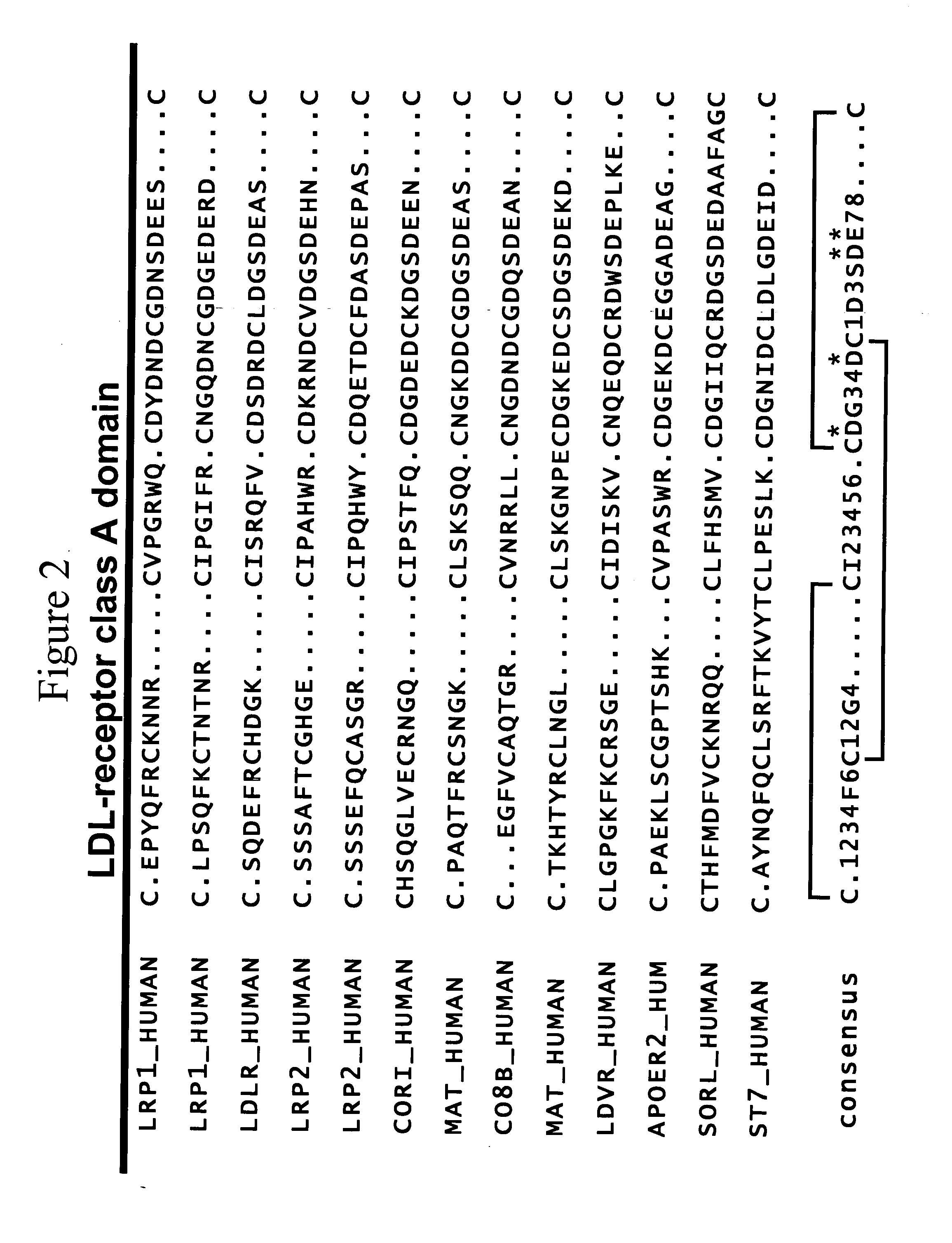
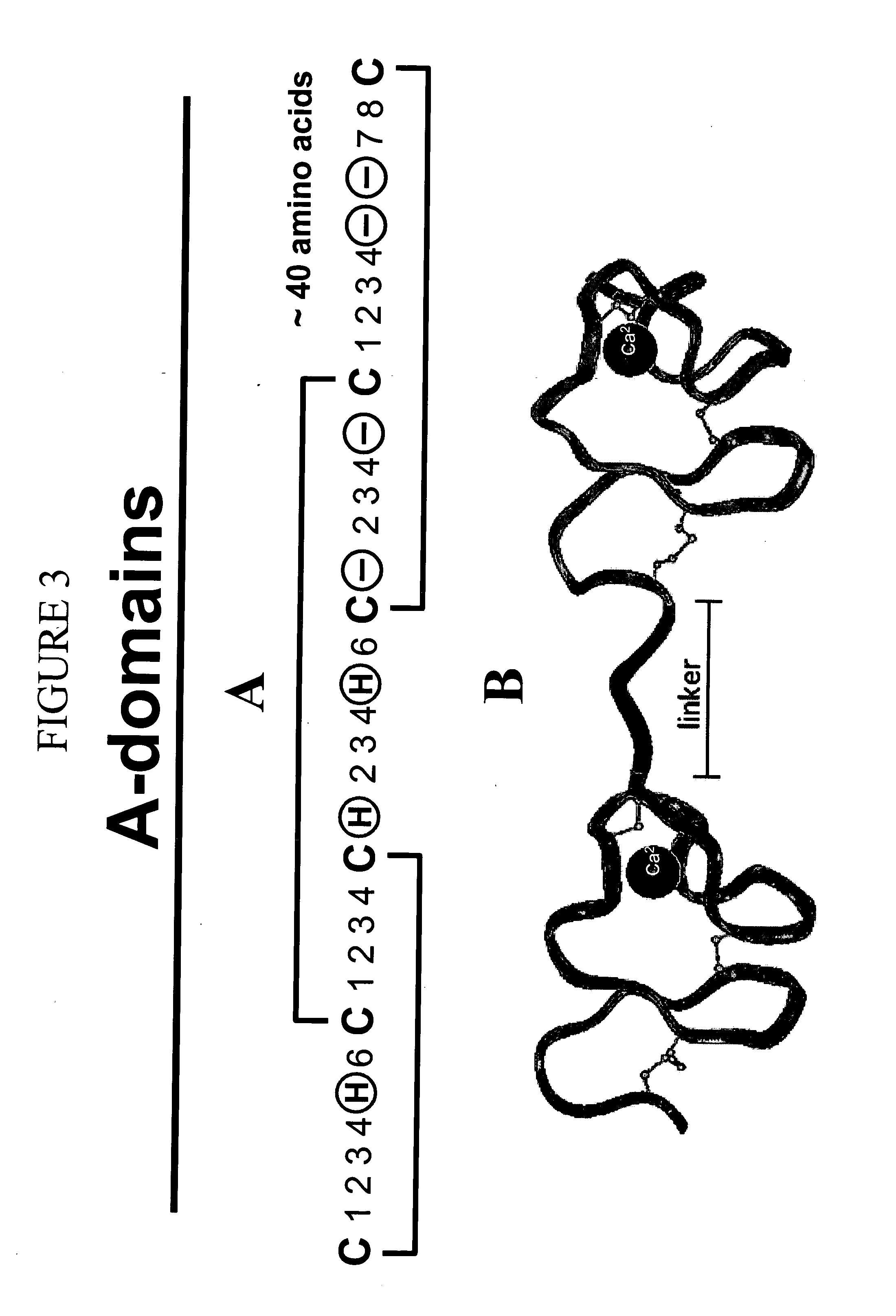
c-Met kinase binding proteins
a technology of kinase and protein, applied in the field of c-met kinase binding proteins, can solve the problems of reducing the tumorigenic potential of human tumour cells, misexpression often correlates with poor prognosis, and downregulation of met or hgf expression in human tumour cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0339] This example describes selection of monomer domains and the creation of multimers.
[0340] Starting materials for identifying monomer domains and creating multimers from the selected monomer domains and procedures can be derived from any of a variety of human and / or non-human sequences. For example, to produce a selected monomer domain with specific binding for a desired ligand or mixture of ligands, one or more monomer domain gene(s) are selected from a family of monomer domains that bind to a certain ligand. The nucleic acid sequences encoding the one or more monomer domain gene can be obtained by PCR amplification of genomic DNA or cDNA, or optionally, can be produced synthetically using overlapping oligonucleotides.
[0341] Most commonly, these sequences are then cloned into a cell surface display format (i.e., bacterial, yeast, or mammalian (COS) cell surface display; phage display) for expression and screening. The recombinant sequences are transfected (transduced or tran...
example 2
[0345] This example describes the selection of monomer domains that are capable of binding to Human Serum Albumin (HSA).
[0346] For the production of phages, E. coli DH10B cells (Invitrogen) were transformed with phage vectors encoding a library of LDL receptor class A-domain variants as a fusions to the pIII phage protein. To transform these cells, the electroporation system MicroPulser (Bio-Rad) was used together with cuvettes provided by the same manufacturer. The DNA solution was mixed with 100 μl of the cell suspension, incubated on ice and transferred into the cuvette (electrode gap 1 mm). After pulsing, 2 ml of SOC medium (2% w / v tryptone, 0.5% w / v yeast extract, 10 mM NaCl, 10 mM MgSO4, 10 mM MgCl2) were added and the transformation mixture was incubated at 37 C for 1 h. Multiple transformations were combined and diluted in 500 ml 2xYT medium containing 201 g / m tetracycline and 2 mM CaCl2. With 10 electroporations using a total of 10 μg ligated DNA 1.2×108 independent clones...
example 3
[0351] This example describes the determination of biological activity of monomer domains that are capable of binding to HSA.
[0352] In order to show the ability of an HSA binding domain to extend the serum half life of a protein in vivo, the following experimental setup was performed. A multimeric A-domain, consisting of an A-domain which was evolved for binding HSA (see Example 2) and a streptavidin binding A-domain was compared to the streptavidin binding A-domain itself. The proteins were injected into mice, which were either loaded or not loaded (as control) with human serum albumin (HSA). Serum levels of a-domain proteins were monitored.
[0353] Therefore, an A-domain, which was evolved for binding HSA (see Example 1) was fused on the genetic level with a streptavidin binding A-domain multimer using standard molecular biology methods (see Maniatis et al.). The resulting genetic construct, coding for an A-domain multimer as well as a hexahistidine tag and a HA tag, were used to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com