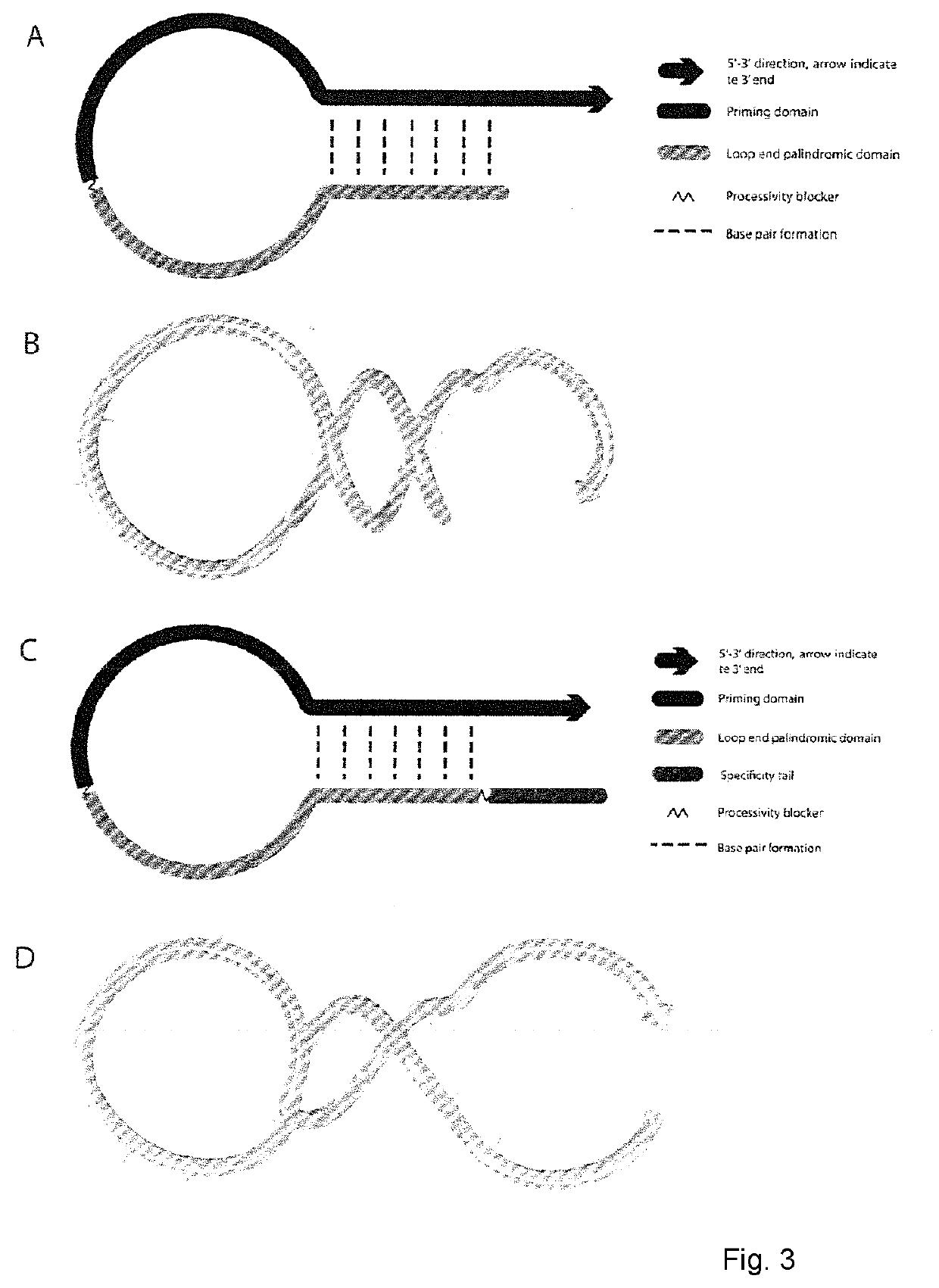
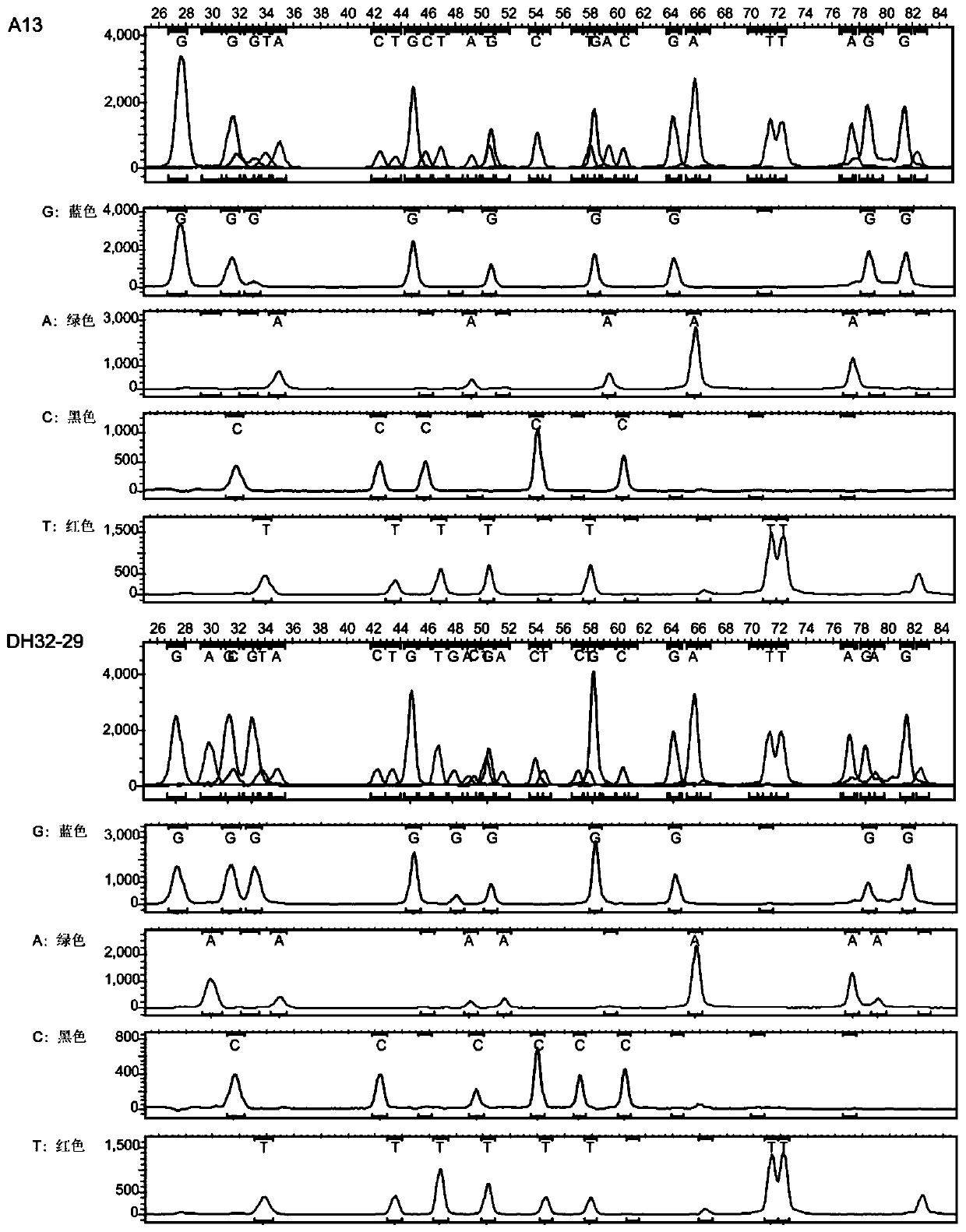
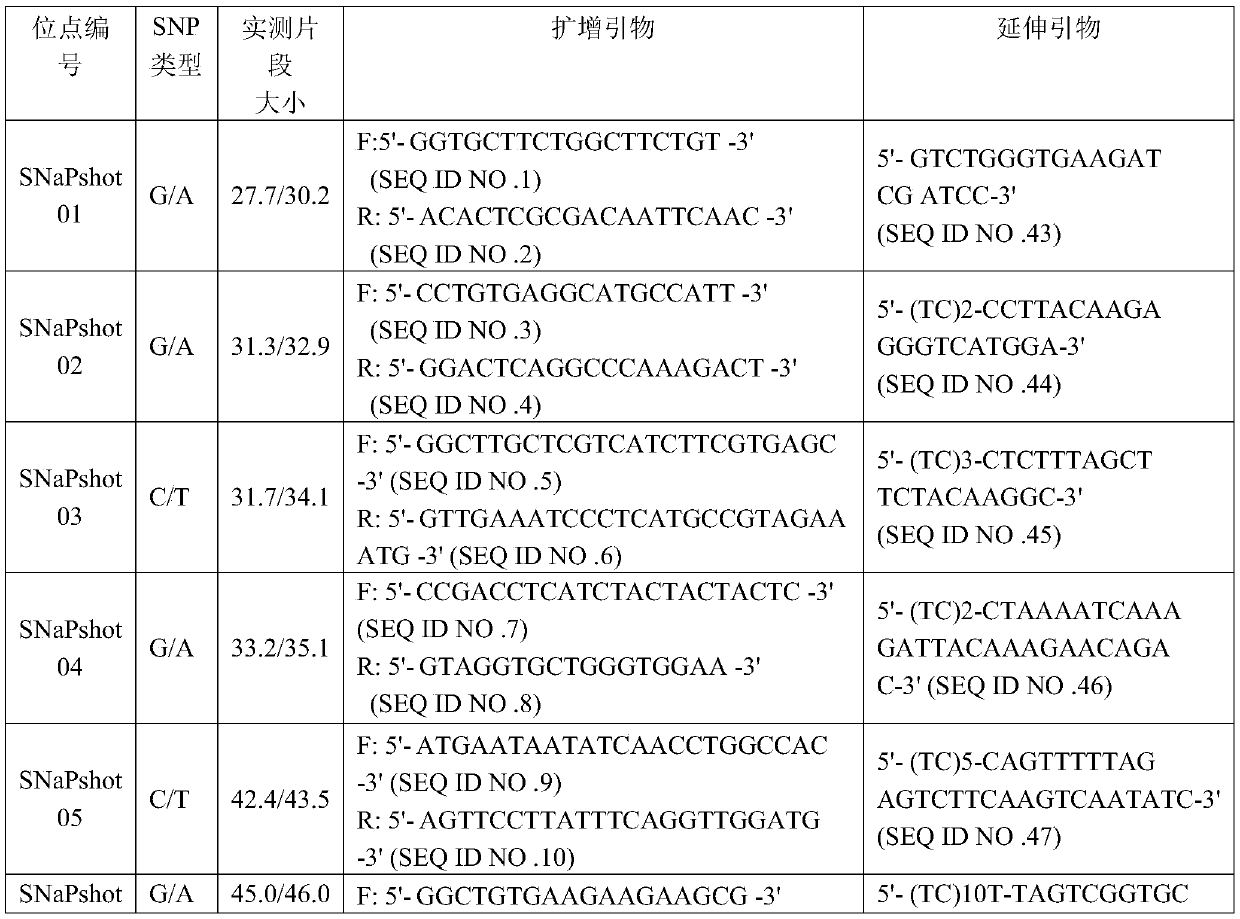
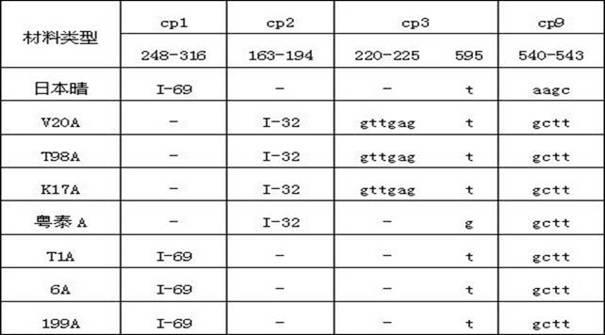
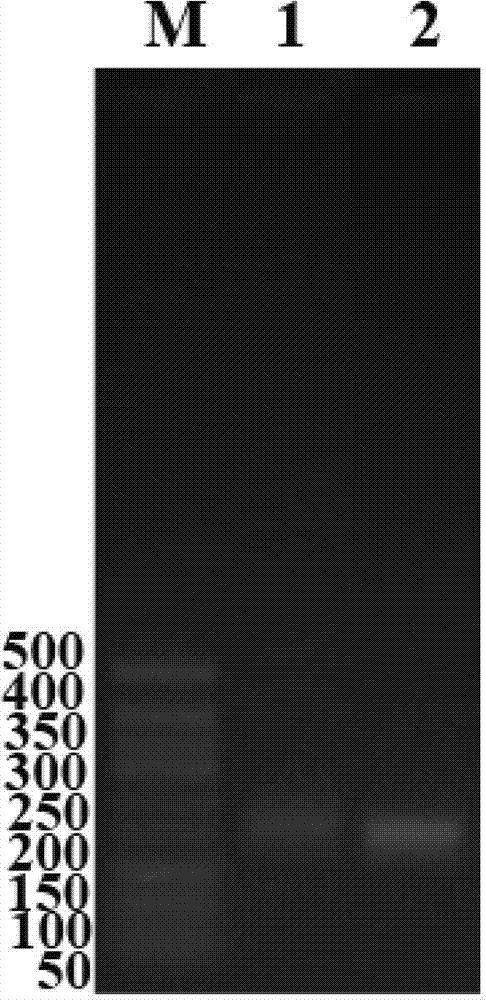
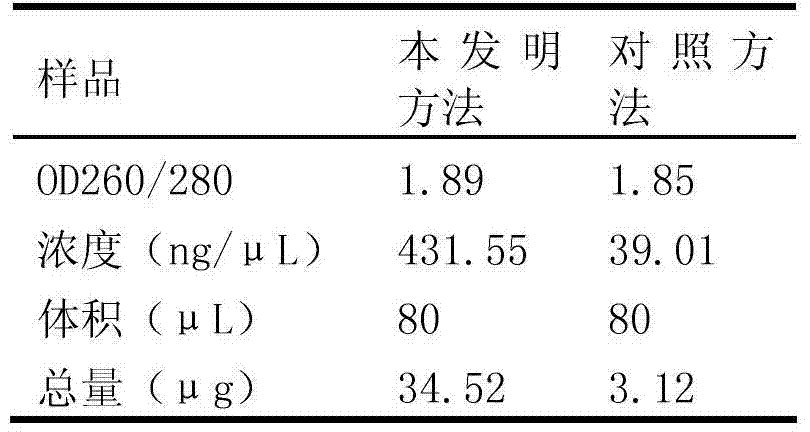
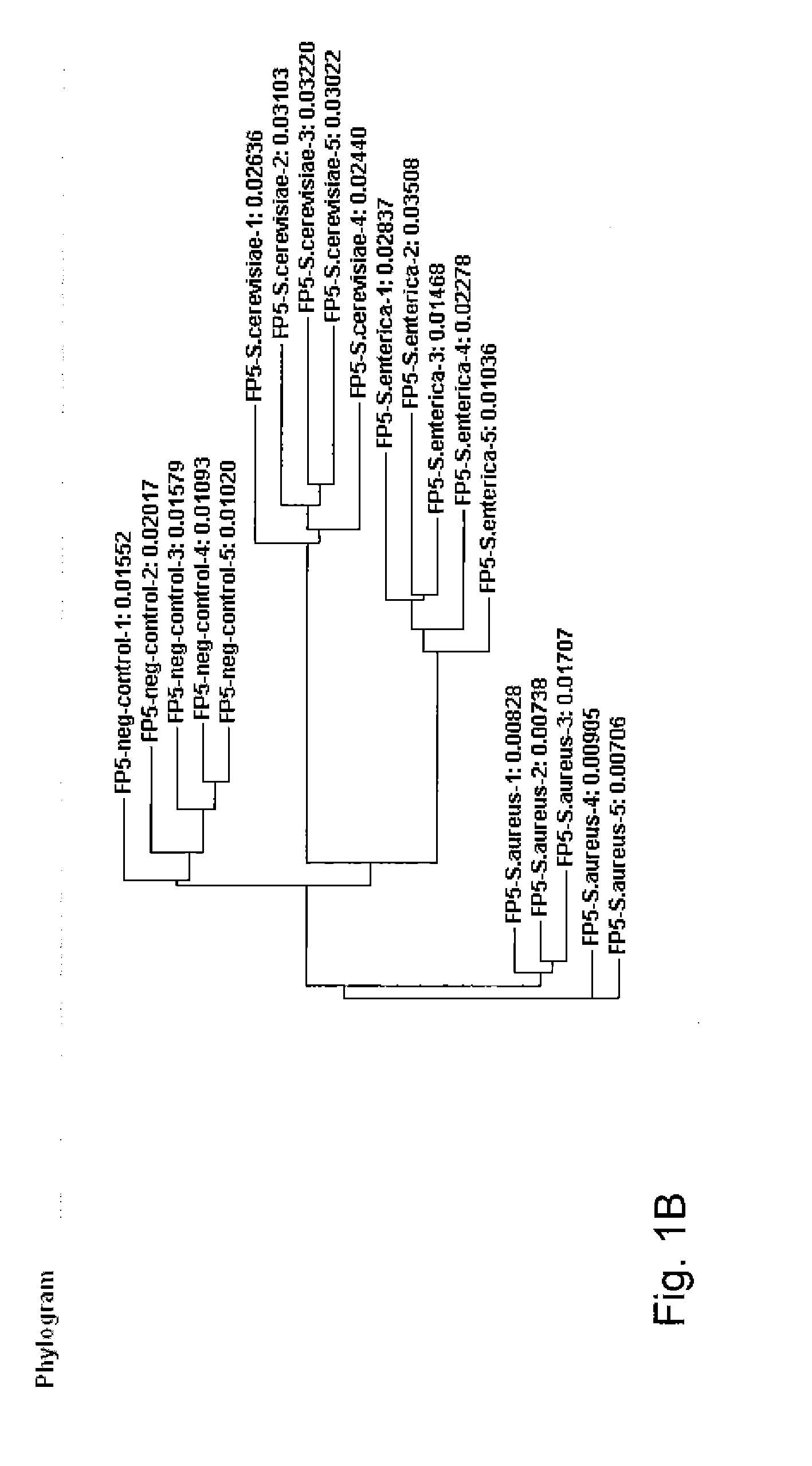
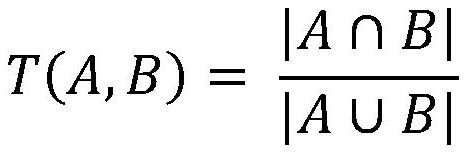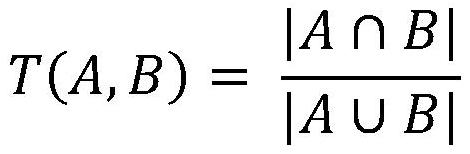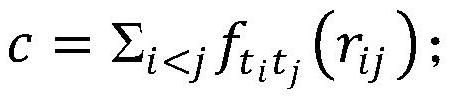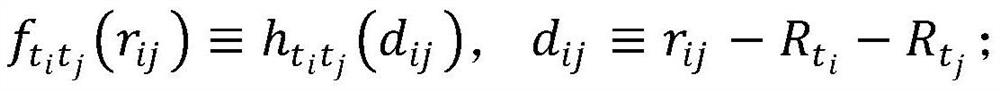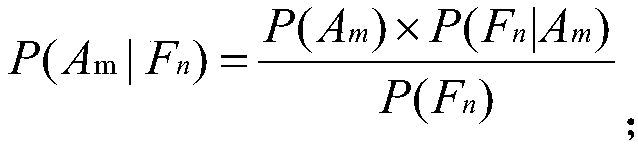Patents
Literature
31 results about "Molecular Fingerprinting" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
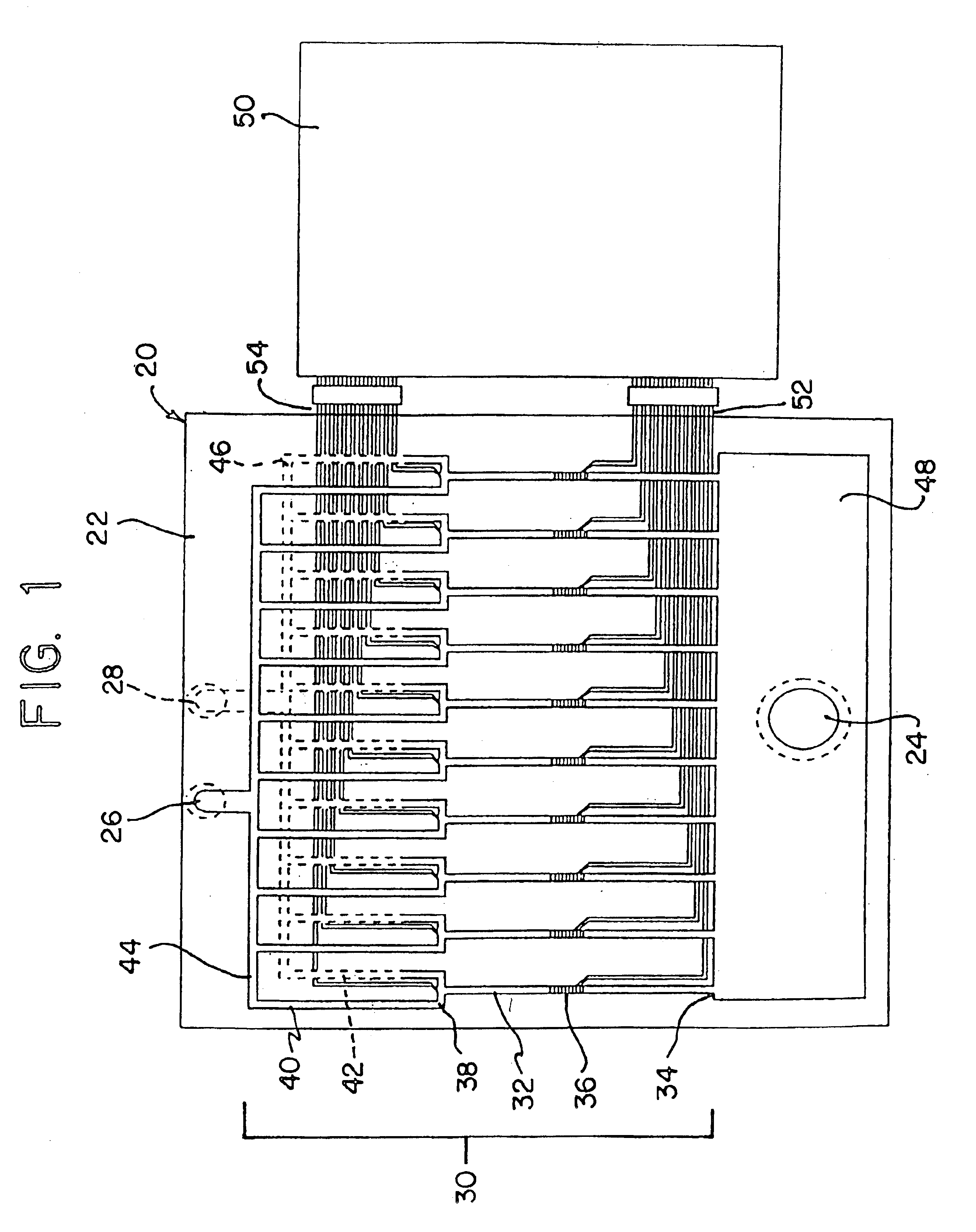
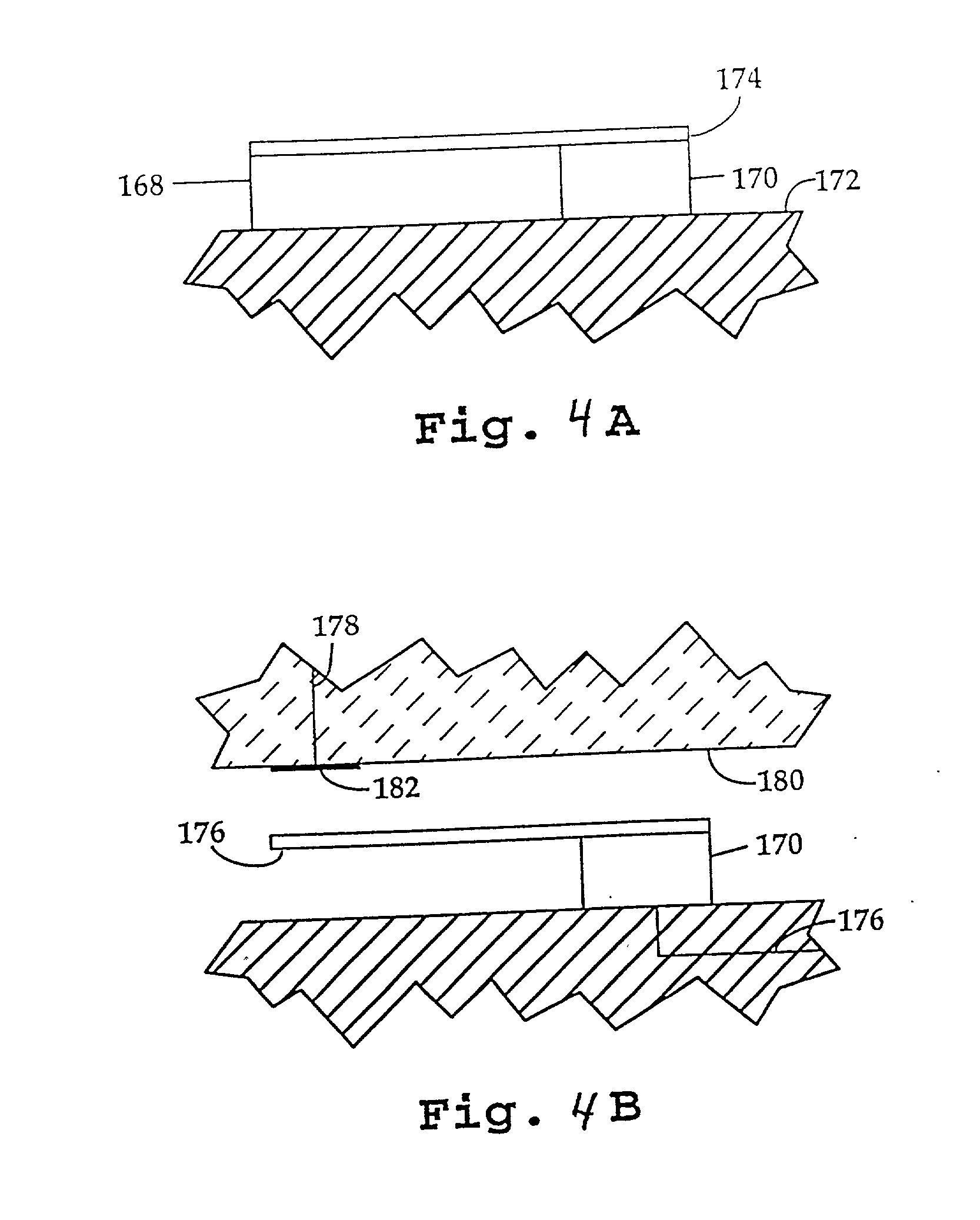
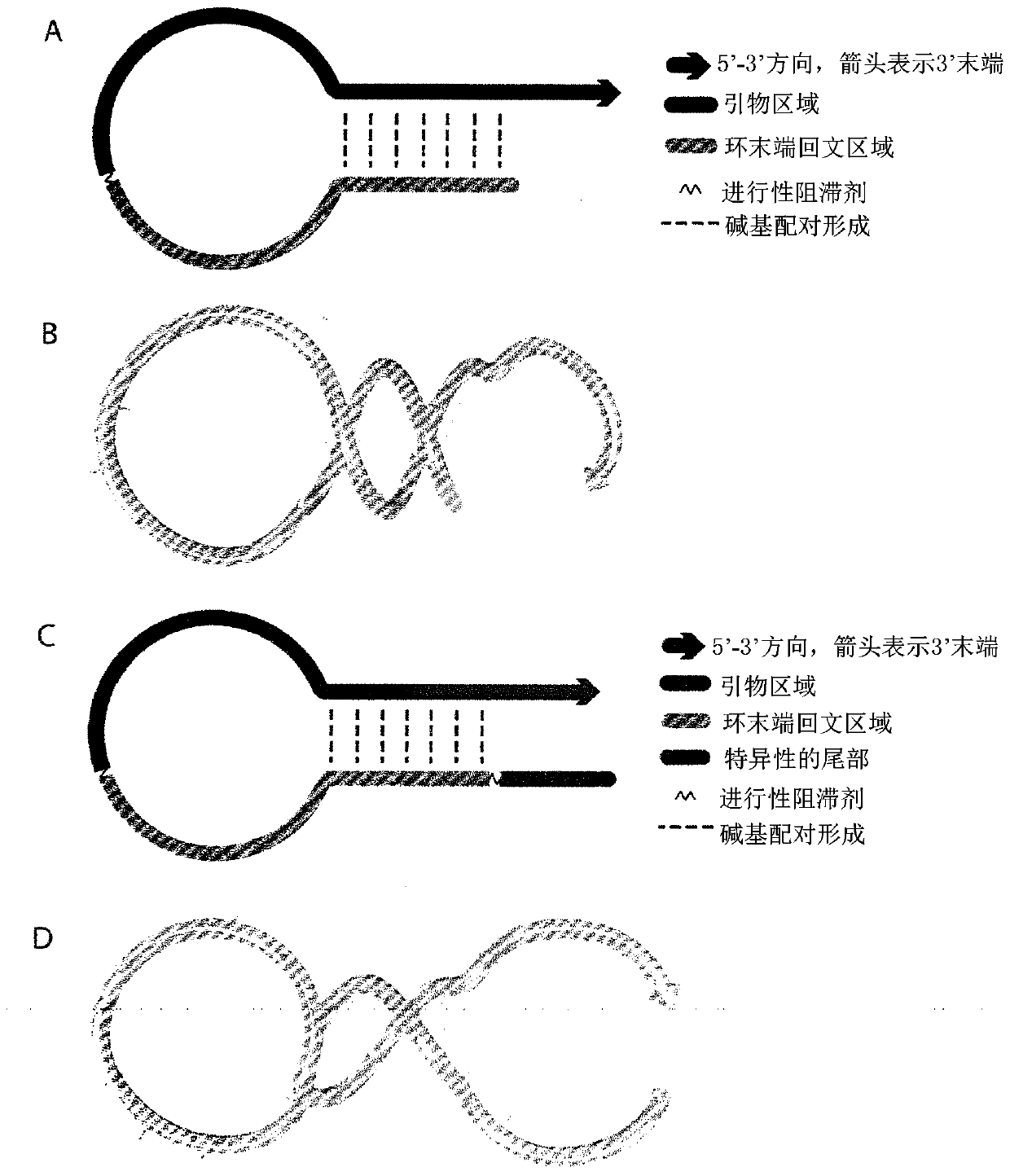
Methods for detecting and sorting polynucleotides based on size
InactiveUS6833242B2Improve discriminationPrevent non-specific bindingBioreactor/fermenter combinationsBiological substance pretreatmentsSmall sampleNucleotide
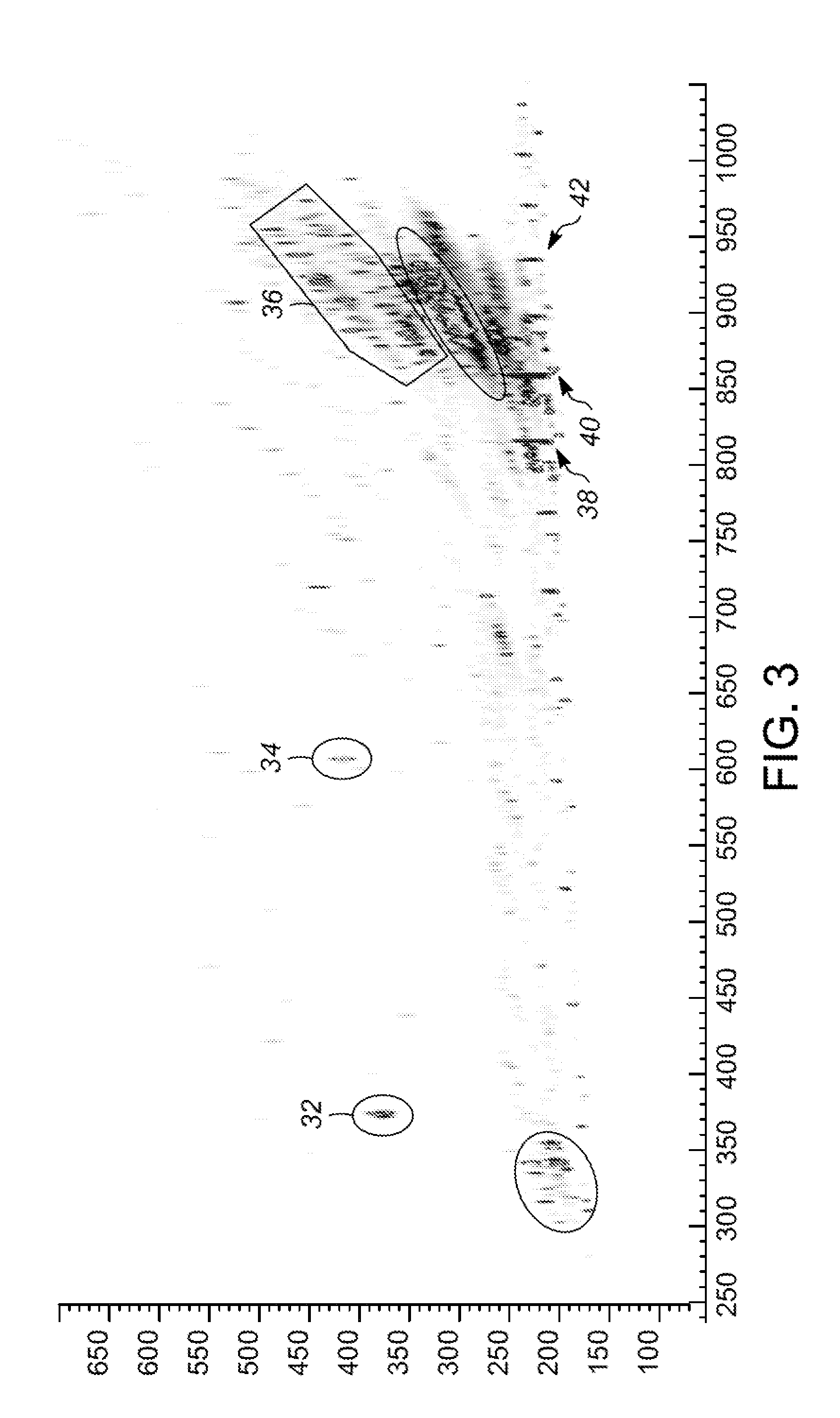
This invention relates in general to a method for molecular fingerprinting. The method can be used for forensic identification (e.g. DNA fingerprinting, especially by VNTR), bacterial typing, and human / animal pathogen diagnosis. More particularly, molecules such as polynucleotides (e.g. DNA) can be assessed or sorted by size in a microfabricated device that analyzes the polynucleotides according to restriction fragment length polymorphism. In a microfabricated device according to the invention, DNA fragments or other molecules can be rapidly and accurately typed using relatively small samples, by measuring for example the signal of an optically-detectable (e.g., fluorescent) reporter associated with the polynucleotide fragments.
Owner:CALIFORNIA INST OF TECH
Remote detection of substance delivery to cells
InactiveUS20050112065A1Highly preventive effectIncreasing effect on proton relaxivityDiagnostics using lightDispersion deliveryLipid formationElectrochemical gradient
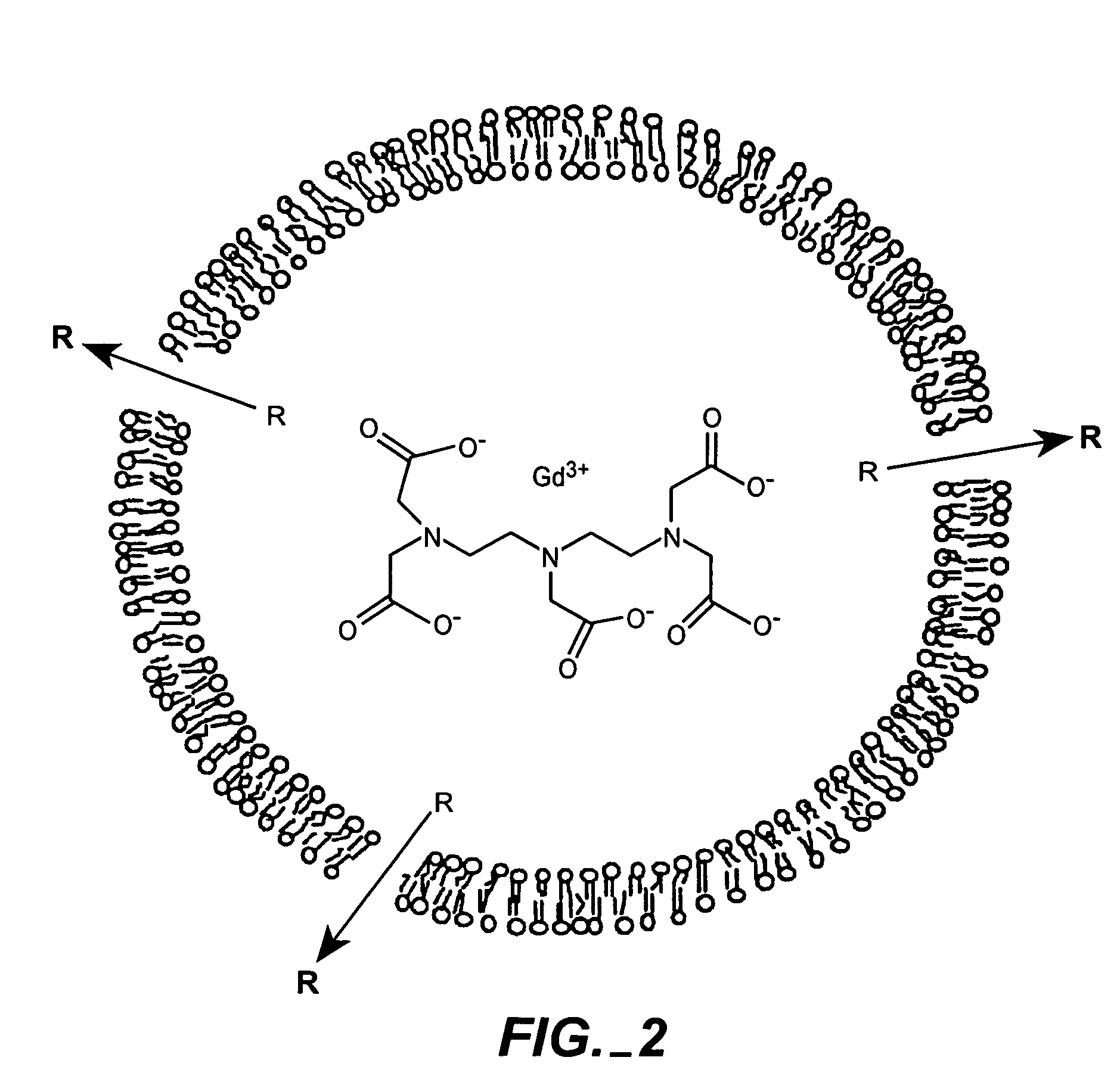
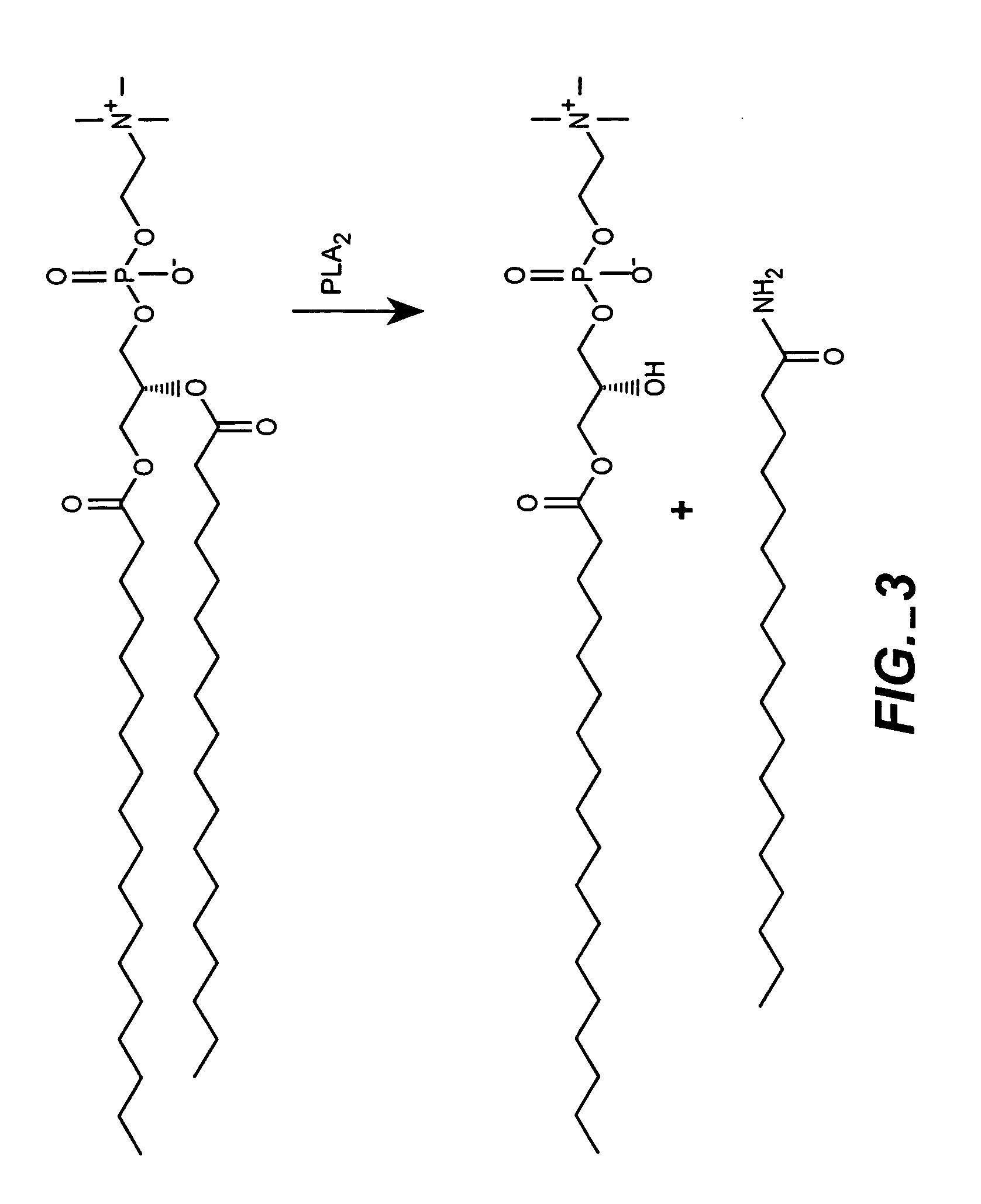
The present invention provides for the development of endocytosis-sensitive probes, and a remote method for measuring cellular endocytosis. These probes are based on the reduced water permeability of a nanoparticle or liposomal delivery system, and inherent degradability or disruption of barrier integrity upon endocytosis. The invention also provides for liposomes having combined therapeutic and diagnostic utilities by co-encapsulating ionically coupled diagnostic and therapeutic agents, in one embodiment, by a method using anionic chelators to prepare electrochemical gradients for loading of amphipathic therapeutic bases into liposomes already encapsulating an imaging agent. The invention provides for imaging of therapeutic liposomes by inserting a lipopolymer anchored, remotely sensing reporter molecules into liposomal lipid layer. The invention allows for an integrated delivery system capable of imaging molecular fingerprints in diseased tissues, treatment, and treatment monitoring.
Owner:SUTTER WEST BAY HOSPITALS
Methods and systems for molecular fingerprinting
InactiveUS20020034748A1Fast and accurate determinationHigh resolutionBioreactor/fermenter combinationsBiological substance pretreatmentsSmall samplePolynucleotide
This invention relates in general to a method for molecular fingerprinting. The method can be used for forensic identification (e.g. DNA fingerprinting, especially by VNTR), bacterial typing, and human / animal pathogen diagnosis. More particularly, molecules such as polynucleotides (e.g. DNA) can be assessed or sorted by size in a microfabricated device that analyzes the polynucleotides according to restriction fragment length polymorphism. In a microfabricated device according to the invention, DNA fragments or other molecules can be rapidly and accurately typed using relatively small samples, by measuring for example the signal of an optically-detectable (e.g., fluorescent) reporter associated with the polynucleotide fragments.
Owner:CALIFORNIA INST OF TECH
Pcr-based genotyping
InactiveUS20110059437A1Improve analysisHigh sample throughputSugar derivativesMicrobiological testing/measurementInsertion sequenceBiology
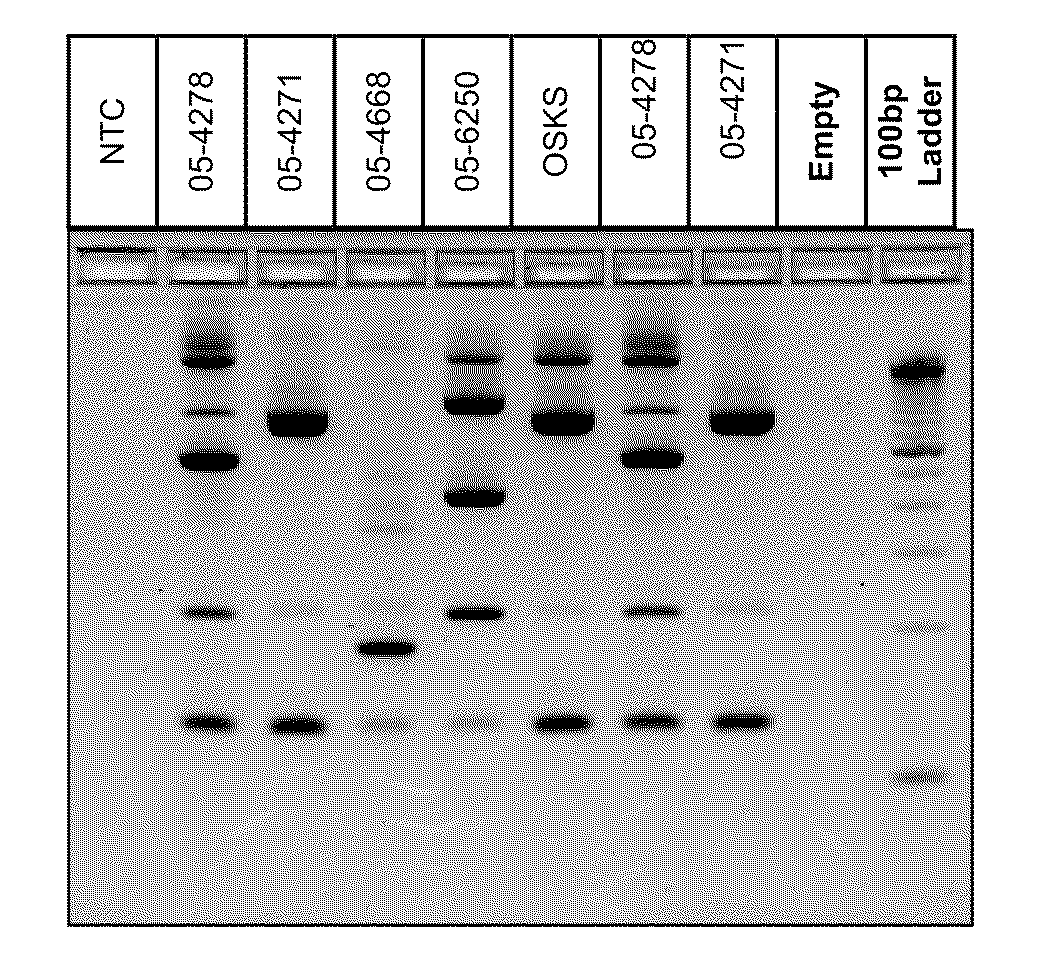
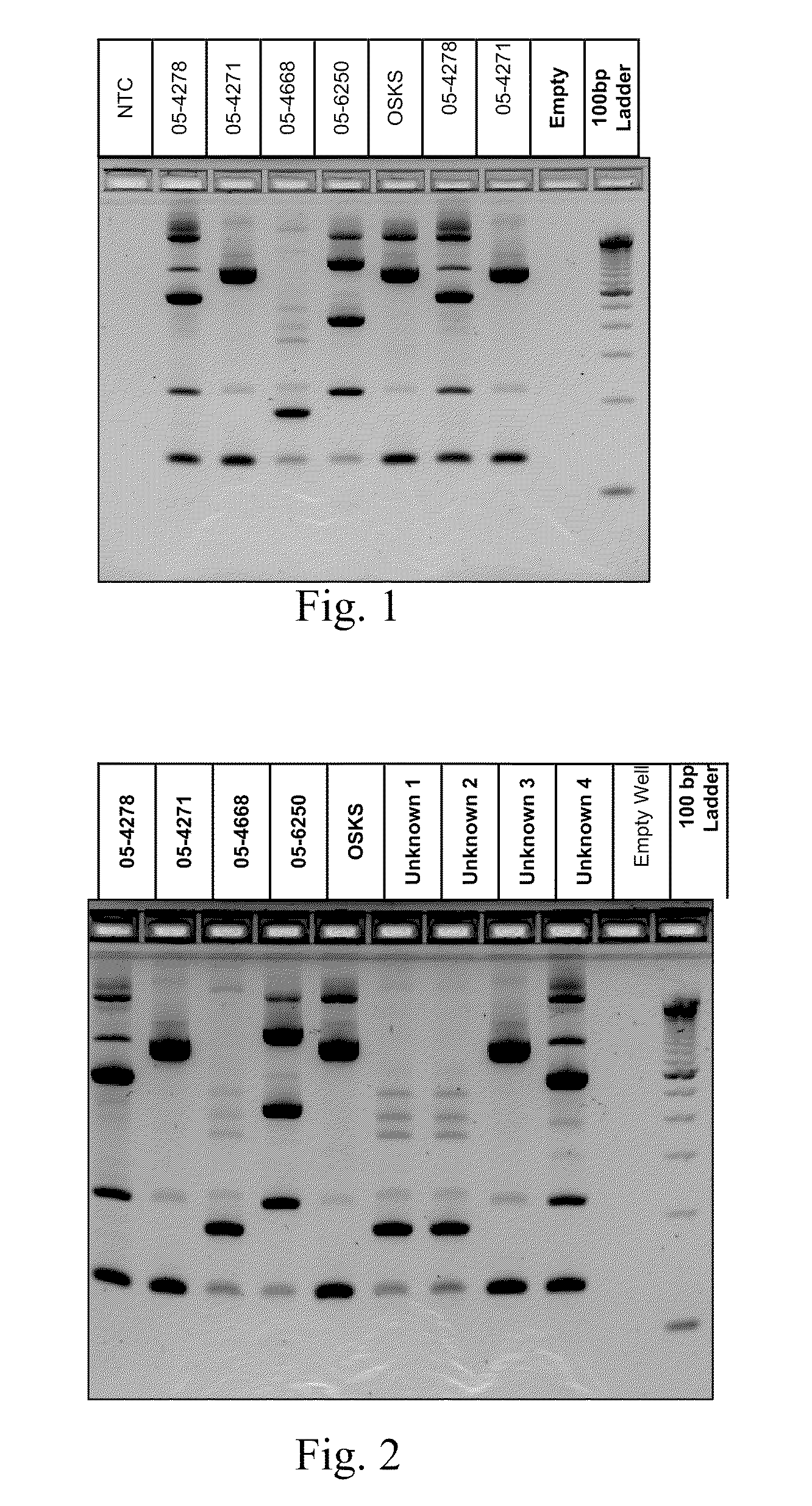
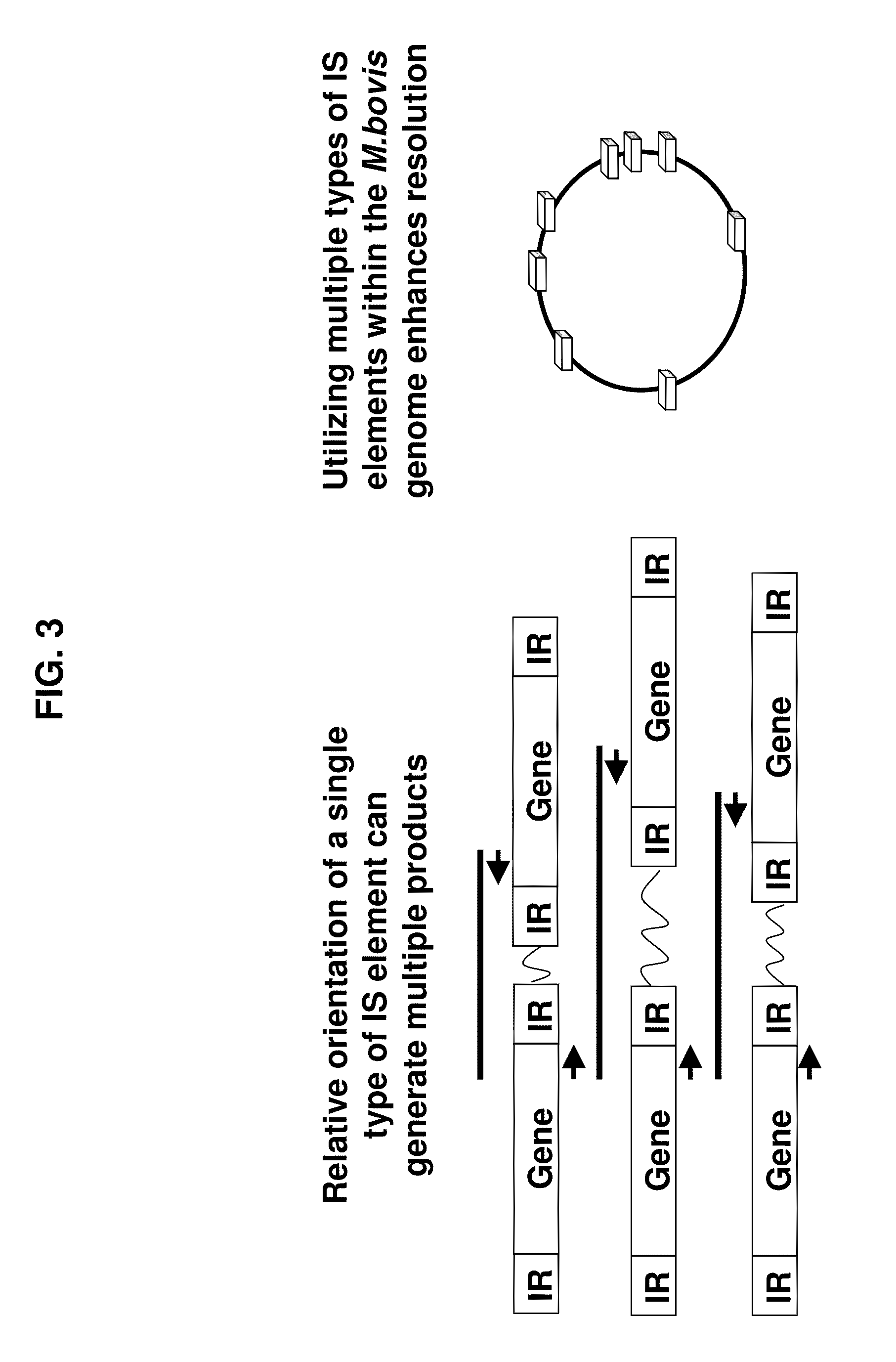
A Mycoplasma bovis PCR-based genotyping method was developed that exploits the proximity of insertion sequences (IS) within the genome by using outward facing primers that selectively amplify sequences between IS elements. The method was applied to 16 field isolates of M. bovis, originating from pneumonic lung or arthritic joints, collected from the United States (Iowa or Kansas) between 2004 and 2005. The genomic fingerprints generated 14 distinct amplification profiles consisting of 4-8 fragments ranging in size from 200-3000 bp. Three isolates presented identical patterns and were isolated from two calves (one calf with pneumonic lung and the other with both pneumonic lung and arthritic joint) from a single farm during an outbreak and probably represent multiple infections with the same genotype. To demonstrate the stability of IS markers for molecular fingerprinting, 3 of the 16 field isolates were subjected to high number passage which resulted in patterns identical to the initial isolates. The results of these studies demonstrate the method can be used for simple and rapid molecular fingerprinting and differentiating M. bovis isolates with extension to epidemiology.
Owner:BOEHRINGER LNGELHEIM VETMEDICA GMBH
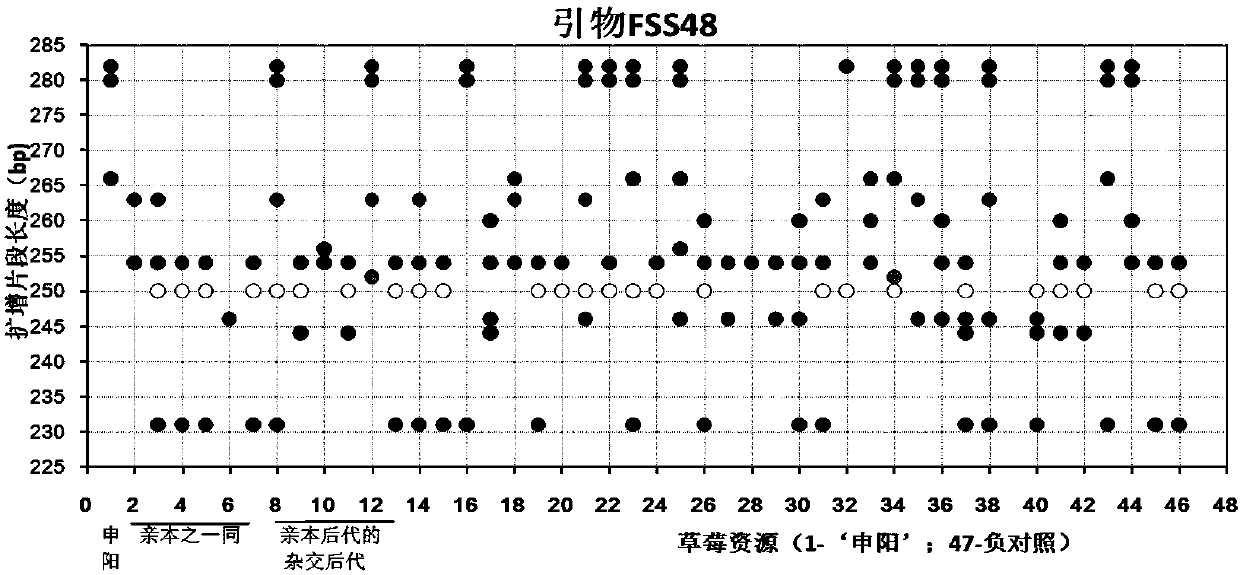
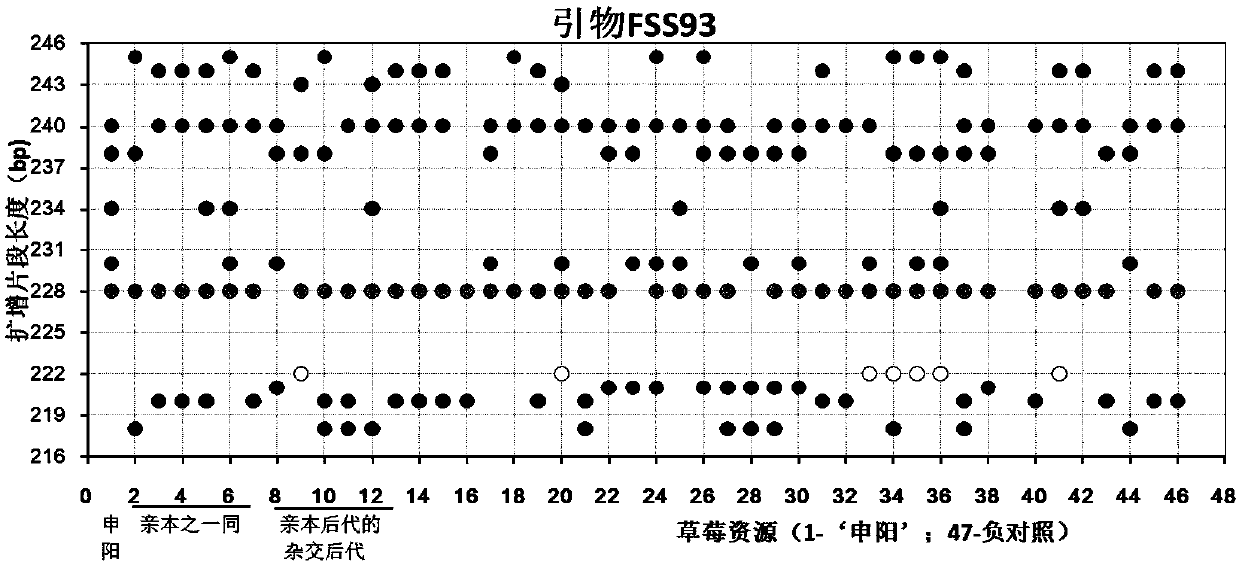
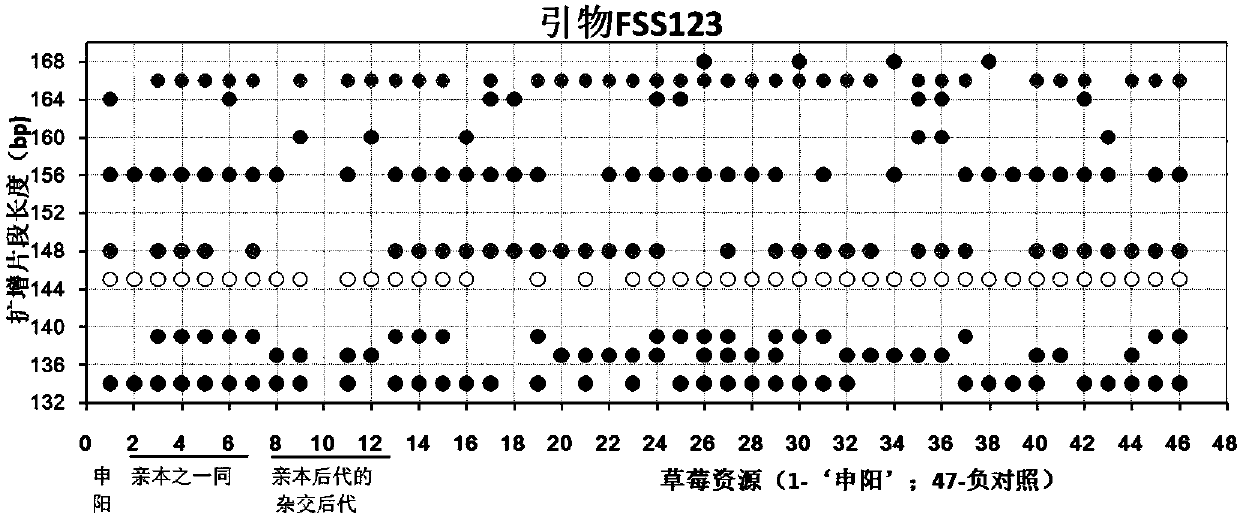
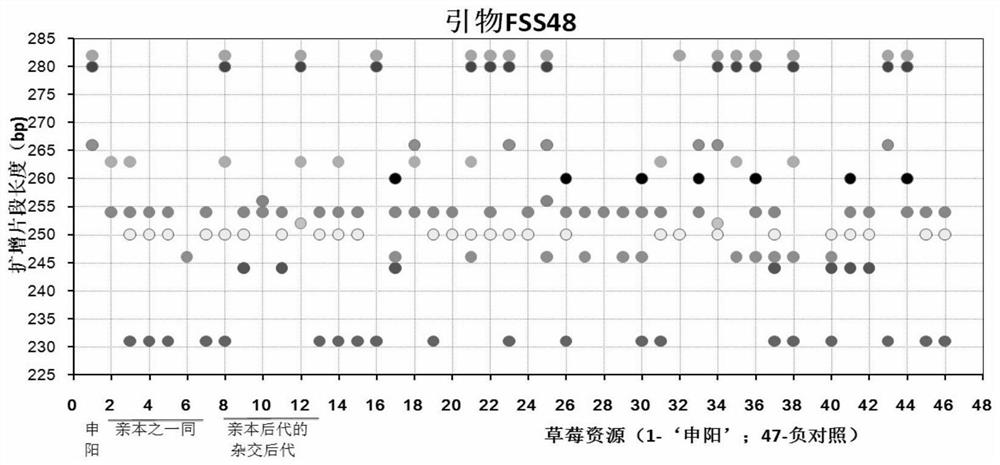
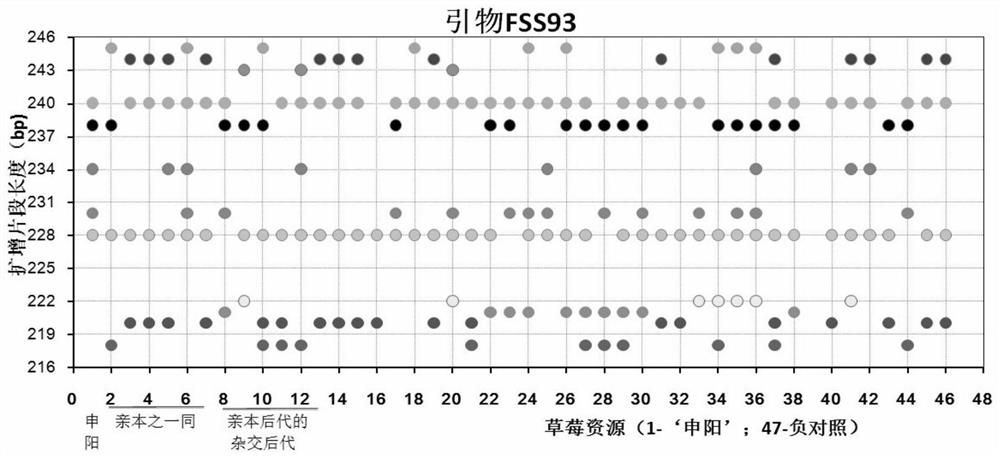
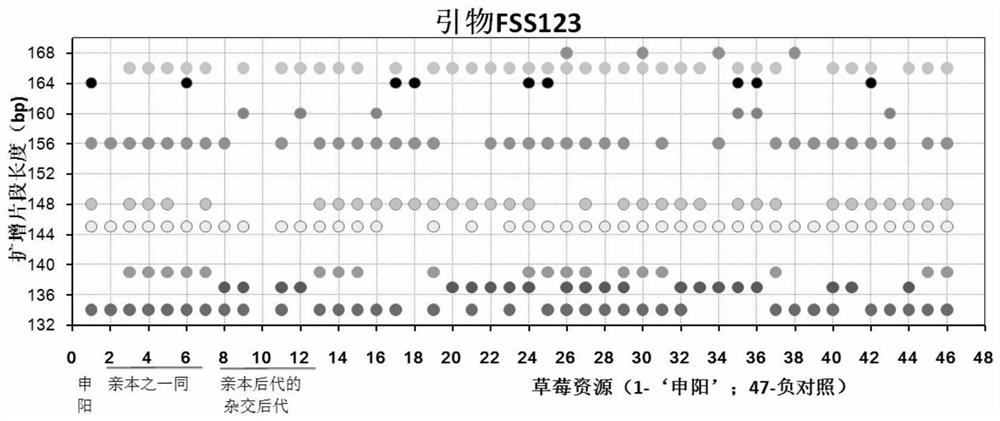
Construction method and application of SSR (Simple Sequence Repeat) molecular fingerprint map of strawberry hybrid
ActiveCN107663548AStrong dominantGood repeatabilityMicrobiological testing/measurementHorticulture methodsGrowth coefficientFragaria
The invention discloses a construction method and application of an SSR (Simple Sequence Repeat) molecular fingerprint map of strawberry hybrid. Eight pairs of SSR primer sequences are screened from 140 pairs of SSR primers which cover the whole genome of strawberries; the SSR molecular fingerprint map of the strawberry hybrid is constructed by utilizing the eight pairs of the SSR primer sequences; the purity of the seedlings of the strawberry hybrid Shenyang, a close relative sib line thereof, a non-near source variety, an excellent line and the like can be authenticated genetically; the SSRmolecular fingerprint map has the characteristics that the specificity is high, the polymorphism is stable, the band type combination is easy to identify and high in dominance, and can authenticate the strawberry hybrid accurately and quickly; moreover, culturing conditions are screened to control a somatic cell mutation risk which is possibly brought by tissue culture; the vitrification and the formation of calluses are stopped; the tissue culture proliferation generation number and the growth coefficient are inhibited; a micropropagation method of the strawberry hybrid Shenyang is determined; the Shenyang strawberry original seedlings with complete varietal purity can be micro-propagated quickly.
Owner:SHANGHAI ACAD OF AGRI SCI
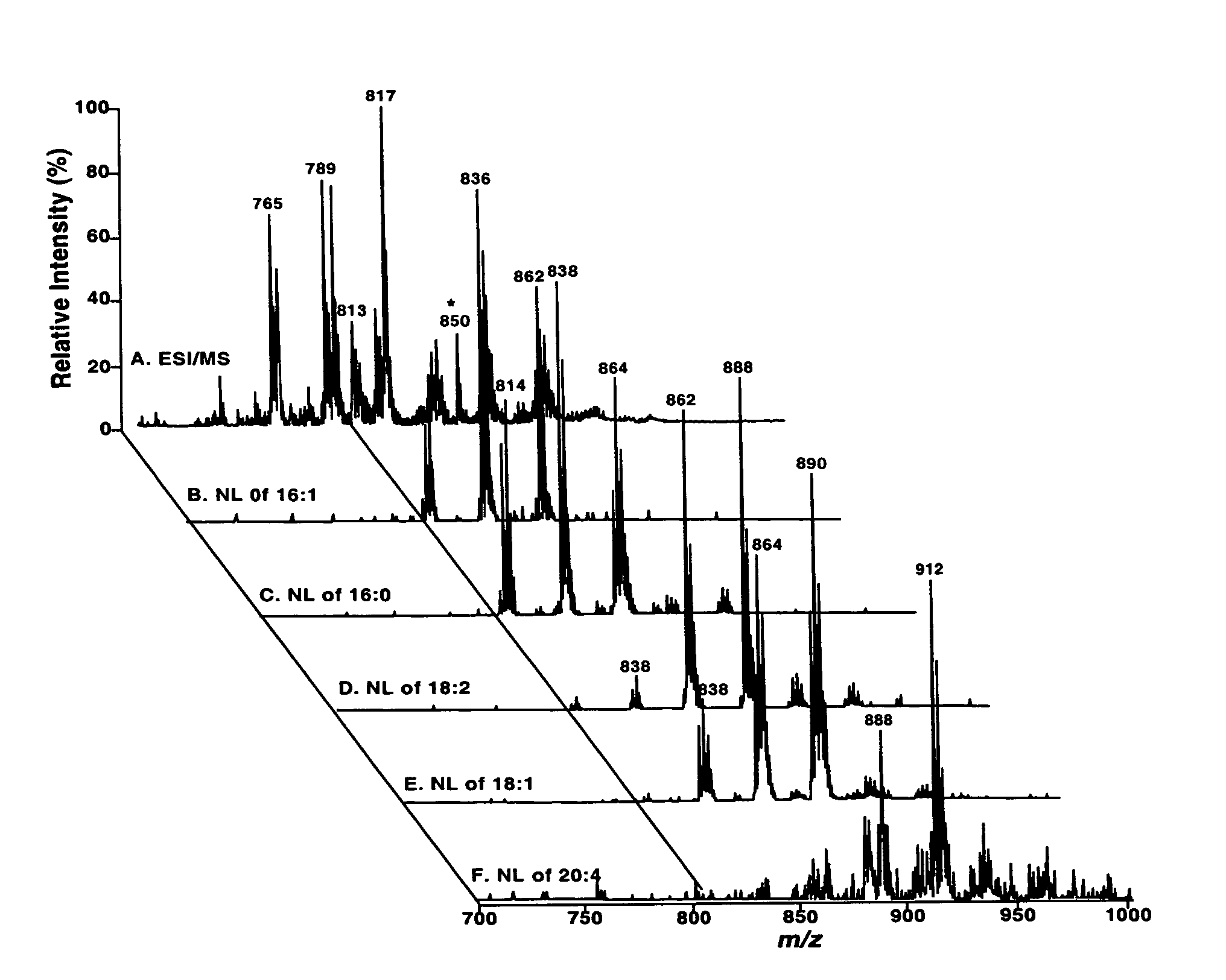
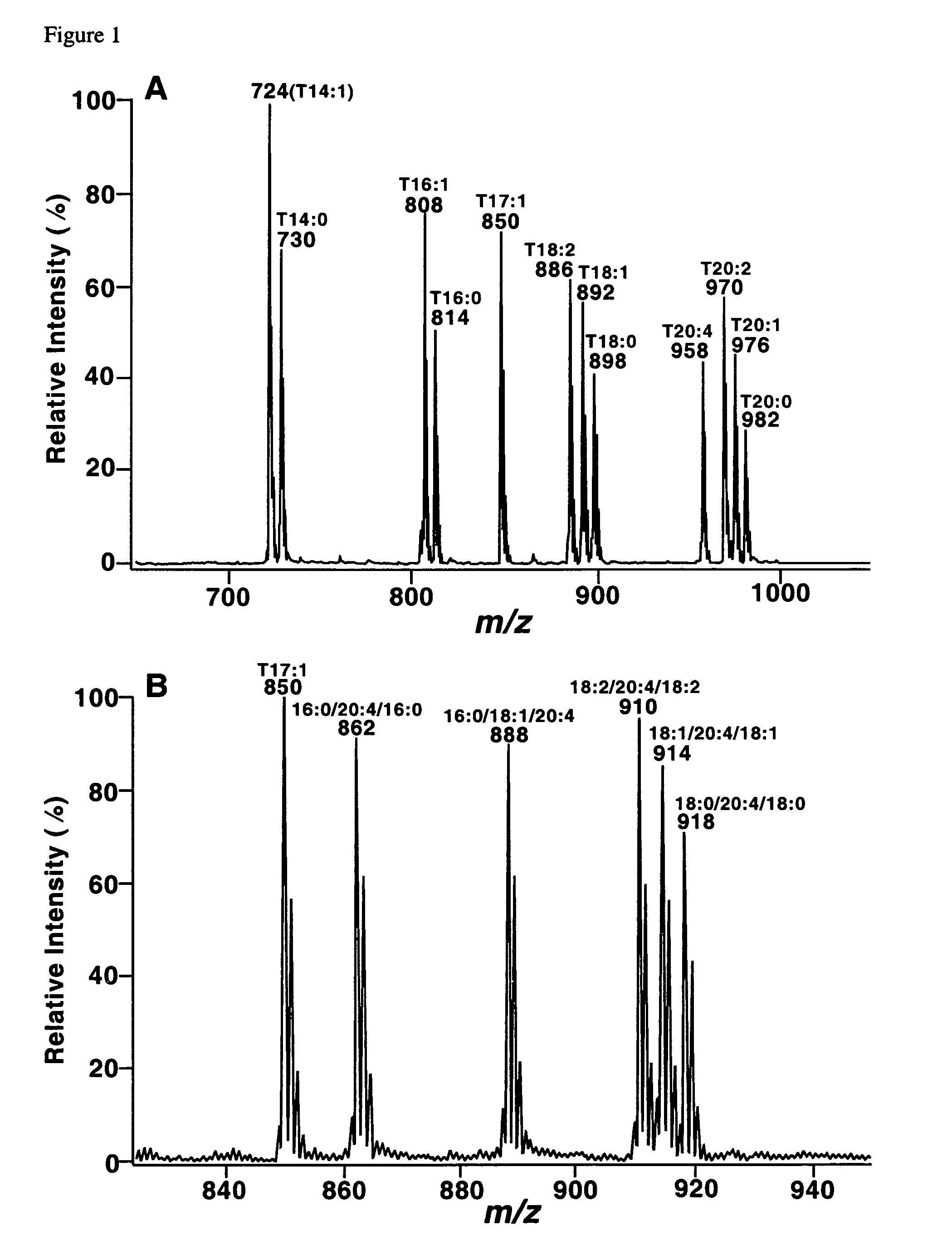
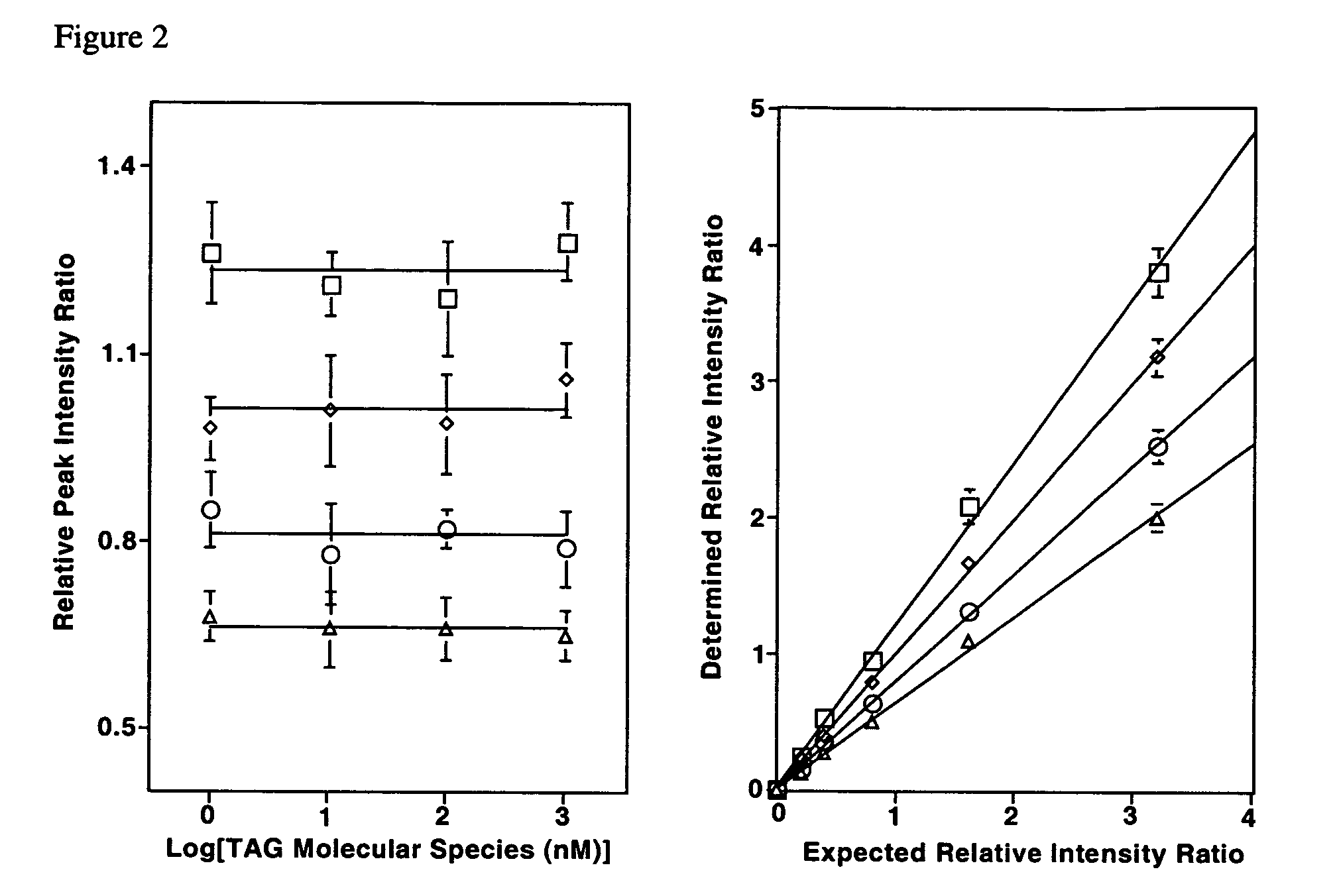
Molecular fingerprinting of triglycerides in biological samples by electrospray ionization tandem mass spectrometry
InactiveUS7306952B2Microbiological testing/measurementFuel testingESI mass spectrometryLipid formation
A method for determination of at least one of the triglyceride molecular species in a biological sample comprising the subjecting sample to lipid extraction to obtain a lipid extract and subjecting the resulting lipid extract to 2D electrospray ionization tandem mass spectrometry (ESI / MS / MS) with neutral loss scanning of all naturally occurring aliphatic chains and contour analysis of 2D intercept peaks. A method for determination of tricylglyceride content and / or TG molecular species directly from a lipid extract of a biological sample comprising subjecting said lipid extract to electrospray ionization tandem mass spectrometry.
Owner:WASHINGTON UNIV IN SAINT LOUIS
Mushroom bacterial standard detecting method
InactiveCN101134979AMicrobiological testing/measurementMaterial analysis by electric/magnetic meansDNA fragmentationMorphological trait
The present invention discloses standard Lentinus edodes seed detecting method, which includes 5 tested aspects including morphological characteristic, agronomic character, physiological character, quality character and molecular fingerprint of special DNA segment, and 36 test indexes, including 34 essential ones and 2 optional ones. Of the 34 essential indexes, there are 10 hypha character indexes and 24 cultivating character indexes.
Owner:上海市农业科学院食用菌研究所
Small molecule drug hERG toxicity prediction method and device based on graph attention mechanism transfer learning
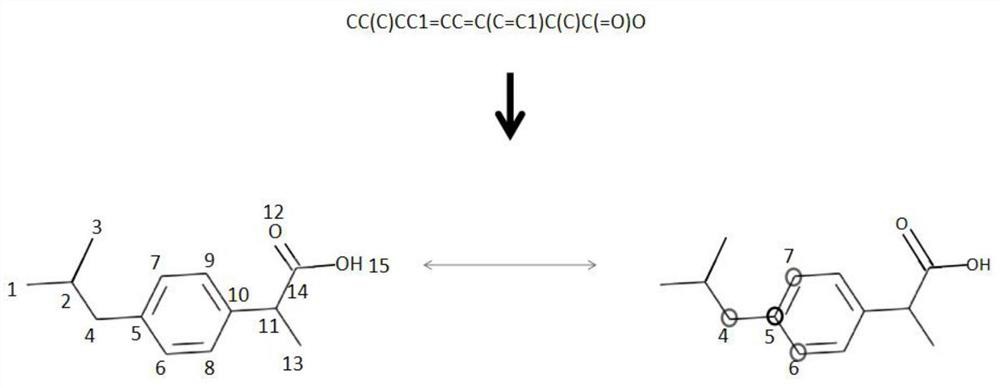
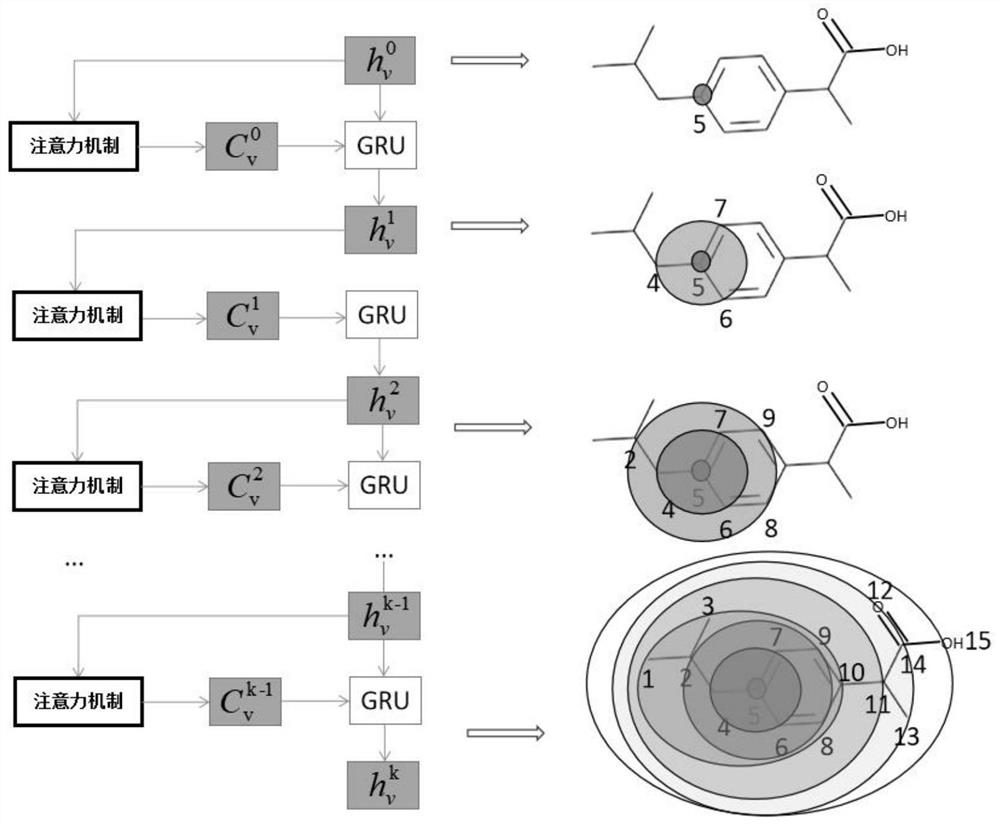
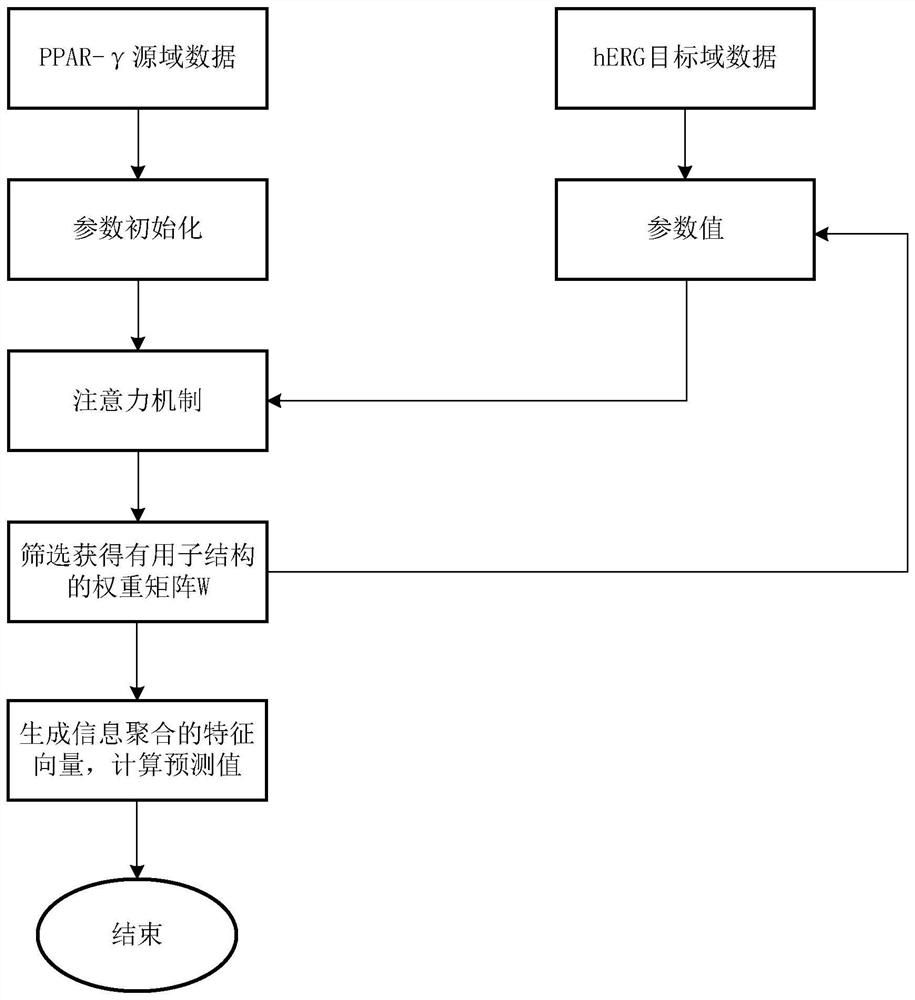
ActiveCN113192571AImprove performancePredictive attribute value contributes a lotChemical property predictionMolecular designData setAlgorithm
The invention discloses a graph attention mechanism transfer learning-based small molecule drug hERG toxicity prediction method and device; the method comprises the following steps: S1, data set preprocessing: enabling a to-be-detected drug-like compound to generate a fingerprint sequence through molecular fingerprint generation software; s2, acquiring atom and chemical bond characteristics through the fingerprint sequence generated in the step S1, and constructing a molecular graph and graph characteristics through the atom and chemical bond characteristics; s3, processing the molecular map obtained in the step S2 through a map attention mechanism, and generating a feature vector of each atom in the molecule; s4, generating a molecular feature vector through a graph attention mechanism and the feature of each atom. According to the method, a molecular graph structure is processed based on a graph attention mechanism, a substructure which contributes to a predicted attribute value is effectively obtained, and a source domain data set and a target domain data set are processed based on transfer learning; and thereby, the problem that the sample size is insufficient is effectively solved.
Owner:NANJING UNIV OF POSTS & TELECOMM
Method for predicting mutagenicity of chemicals through machine learning algorithm
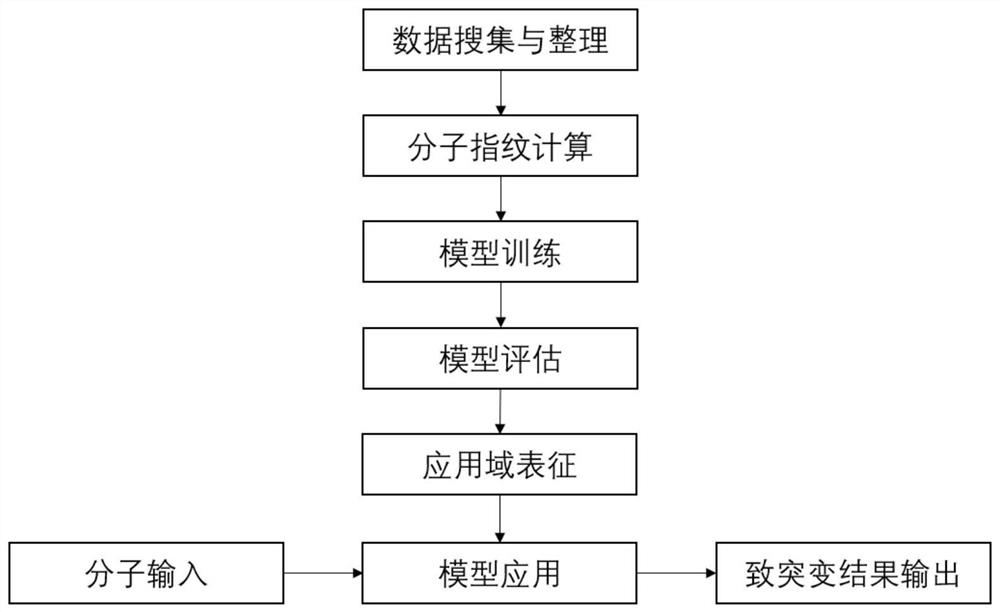
ActiveCN112466399AThe method is simple and fastLow costCharacter and pattern recognitionProteomicsExperimental testingAlgorithm
The invention belongs to the field of ecological risk evaluation test strategies, and discloses a method for predicting mutagenicity of chemicals through a machine learning algorithm. On the basis ofthe known compound molecular structure, the mutagenicity of the compound can be quickly and efficiently predicted by calculating the molecular fingerprint and applying the constructed method. The method is simple and quick, is low in cost, and can save manpower, material resources and financial resources required by experiment testing. The construction process of the method comprises the followingsteps: collecting chemical mutagenicity data; preprocessing the data; calculating a molecular fingerprint; selecting a machine learning algorithm and training a model; selecting indexes such as accuracy to evaluate the model; characterizing the application domain; in the construction method, inputting molecules to be detected, and outputting mutagenicity of the molecules to be detected. The prediction model established by the method has good fitting capability, robustness and prediction capability, can effectively predict mutagenicity of the chemicals, provides necessary basic data for risk evaluation and management of the chemicals, and has important significance.
Owner:DALIAN UNIV OF TECH
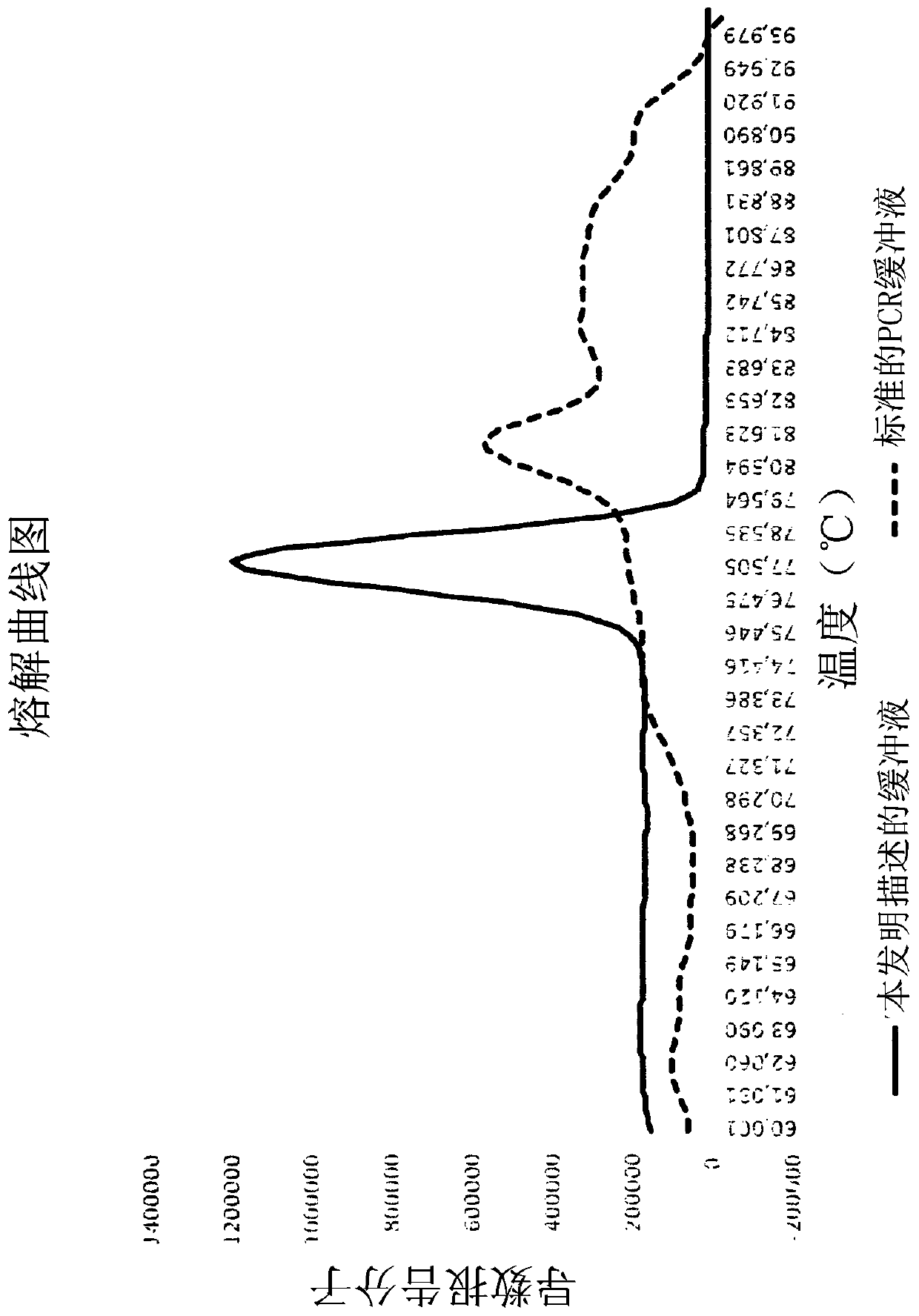
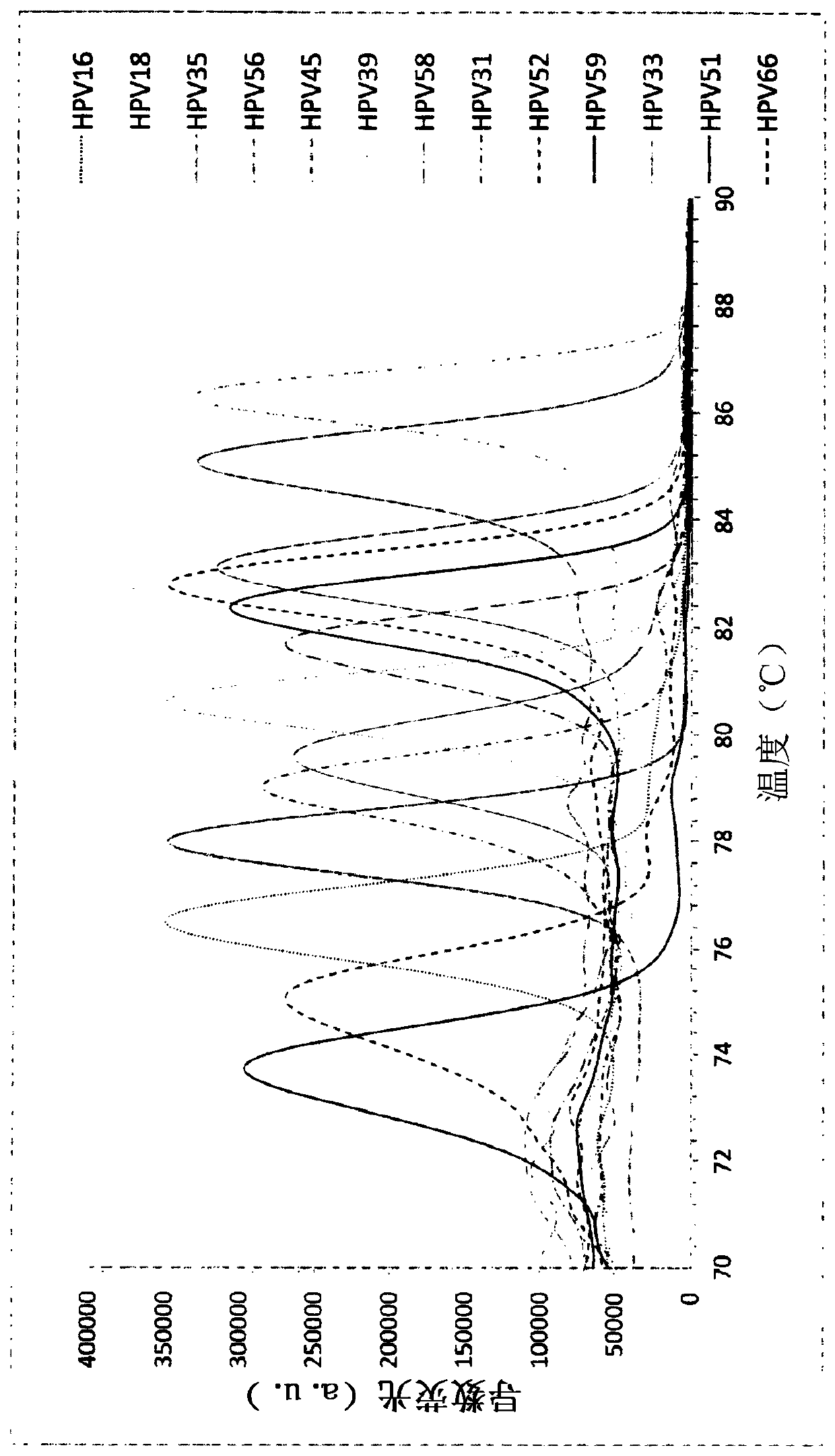
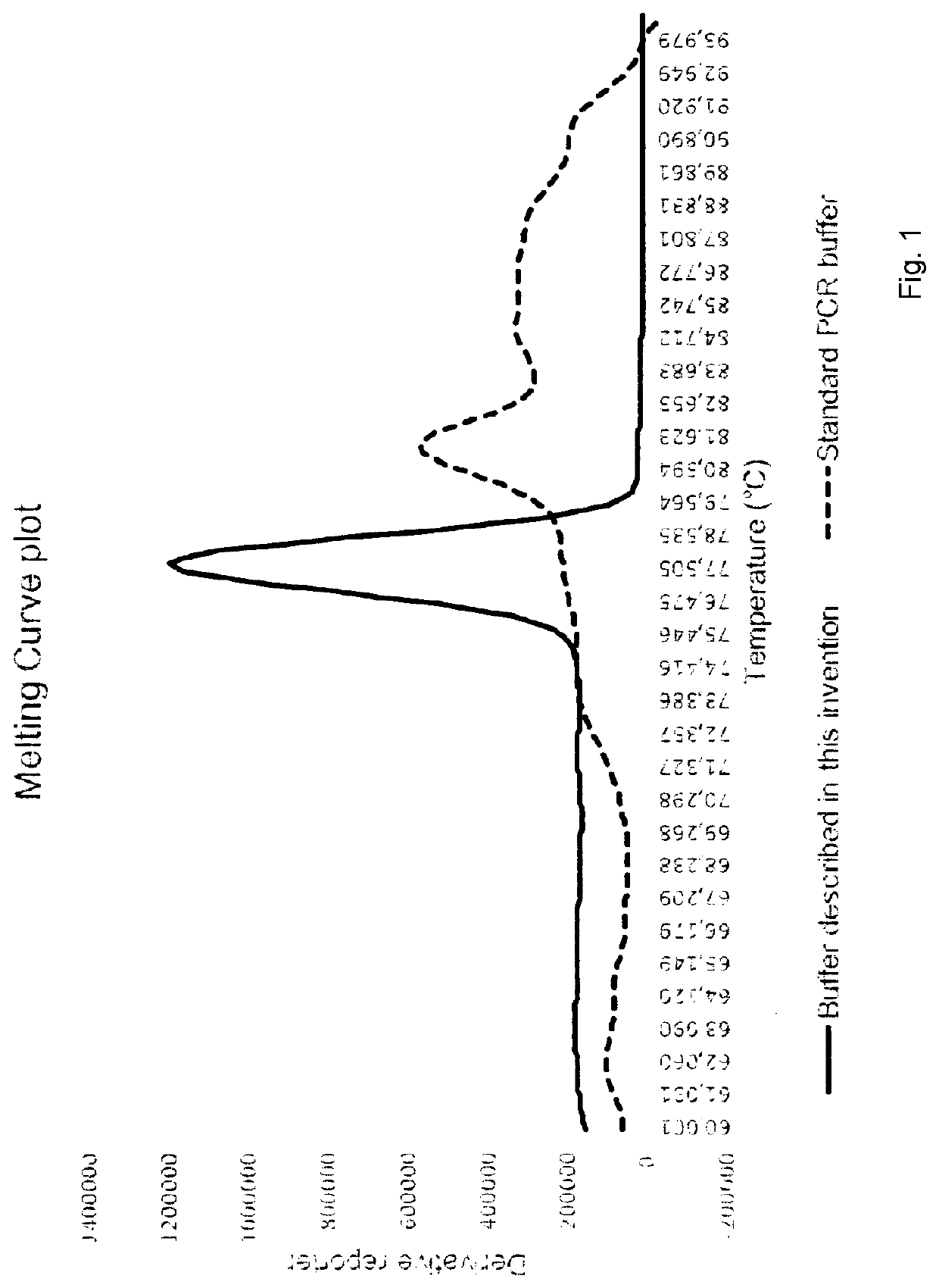
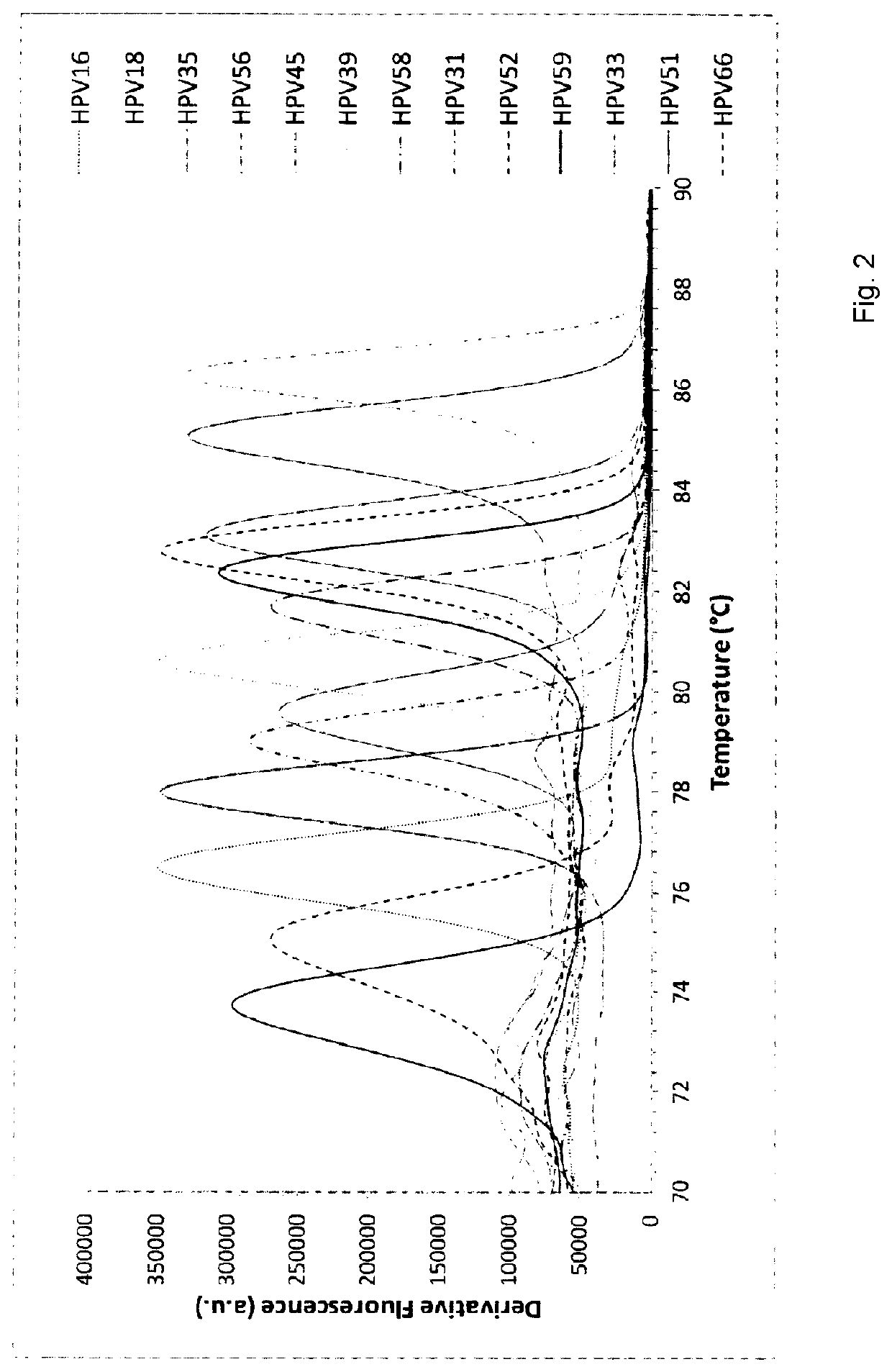
Molecular fingerprinting methods to detect and genotype dna targets through polymerase chain reaction
A molecular fingerprinting method is disclosed to detect and genotype pathogens DNA targets in a sample through polymerase chain reaction (PCR). The method comprises performing PCR amplification and then a High Resolution Melting (HRM) analysis. The PCR reaction mixture used comprises two or more pairs of amplification primers designed in order to generate amplicons with a different melting temperature each other in order to discriminate, in the HRM analysis, each amplicon by observing the specific melting temperature of each amplicon. The method further comprises monitoring, during the HRM analysis, the change in the signal emission resulting from the temperature-induced denaturation of the double-stranded amplicons into two single- stranded DNAs, due to the release of the intercalating molecule or compound. Discrimination and genotyping of different strains of the same pathogen, different pathogens belonging to separated genus and genetic variations in the sample can be determined through a reader analysing the signal variation; the result of the analysis is obtained through a graphic interface connected to the reader.
Owner:ULISSE BIOMED
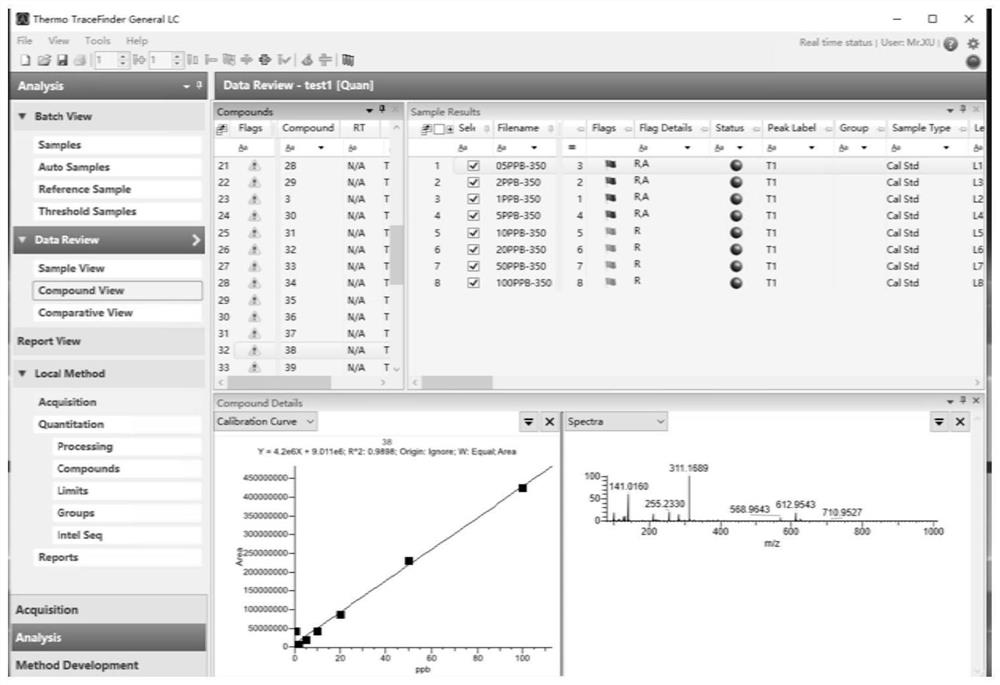
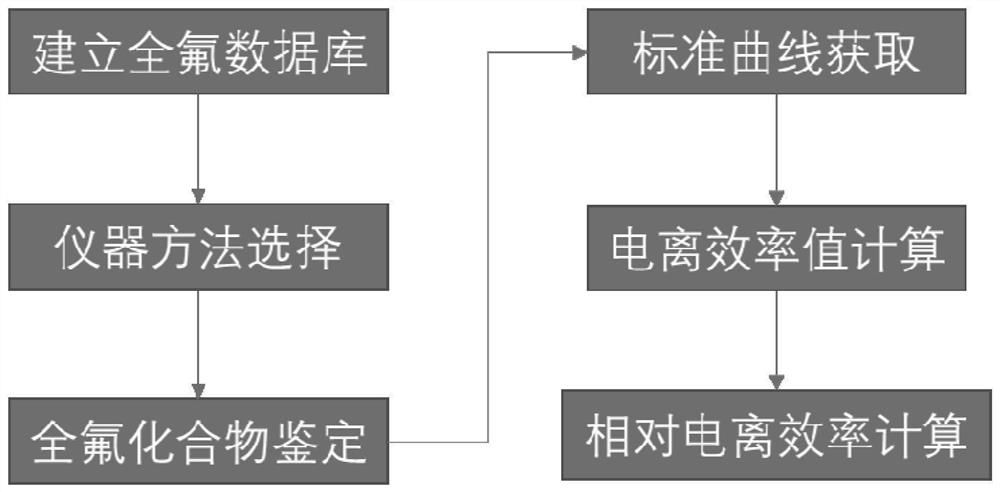
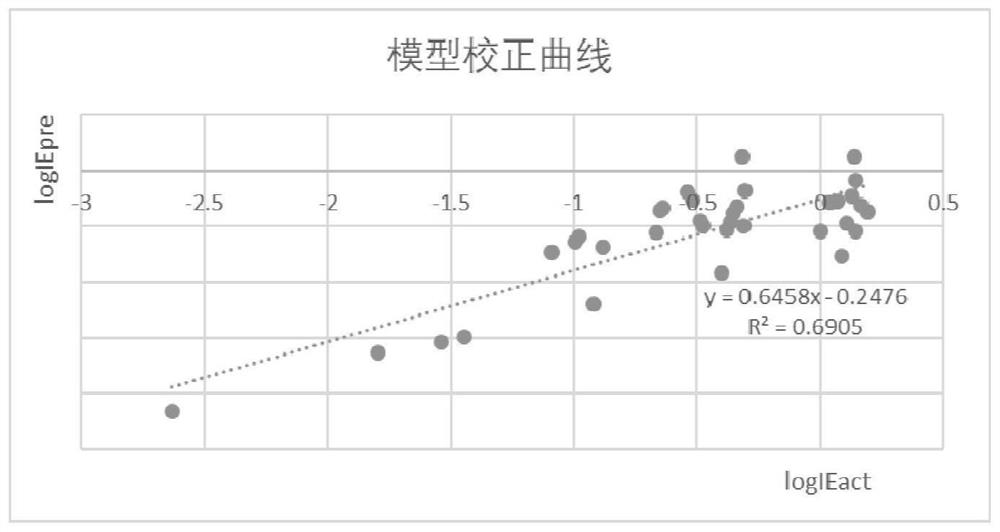
Perfluorinated compound non-targeted quantification method based on ultra-high performance liquid chromatography and high-resolution mass spectrometry
PendingCN114414689AQuantitatively accurateGood explanatoryComponent separationFluid phaseResolution (mass spectrometry)
The invention discloses a perfluorinated compound Chinese chive targeted quantification method based on ultra-high performance liquid chromatography and high-resolution mass spectrometry, and belongs to the field of analytical chemistry. The method comprises the following steps: pre-treating a sample; detecting on a machine: determining the sample through ultra-high performance liquid chromatography-quadrupole orbitrap mass spectrometry; determining the ionization efficiency value of the perfluorinated compound; the structural characterization method of the perfluorinated compound comprises the following steps: molecular fingerprints are obtained; machine learning modeling and evaluation: establishing a machine learning model of molecular fingerprint prediction ionization efficiency; the application of the model comprises the following steps: adding a small amount of different standard substances into an actual sample, detecting the peak area of the standard substances and the response factor RF of the ratio of the peak area to the concentration, fitting a logRF and ionization efficiency curve, and predicting the ionization efficiency value and the unknown substance peak area according to the model to quantify the concentration in a non-targeted manner. According to the method, model correction is carried out through a small amount of standard substances of an actual sample, the method is applied to actual environmental samples, and non-targeted quantification of perfluorinated compounds can be achieved.
Owner:NANJING UNIV
Construction method and application of ssr molecular fingerprint of "Shenyang" strawberry
ActiveCN107663548BStrong specificityStrong discriminationMicrobiological testing/measurementHorticulture methodsBiotechnologyFragaria
Owner:SHANGHAI ACAD OF AGRI SCI
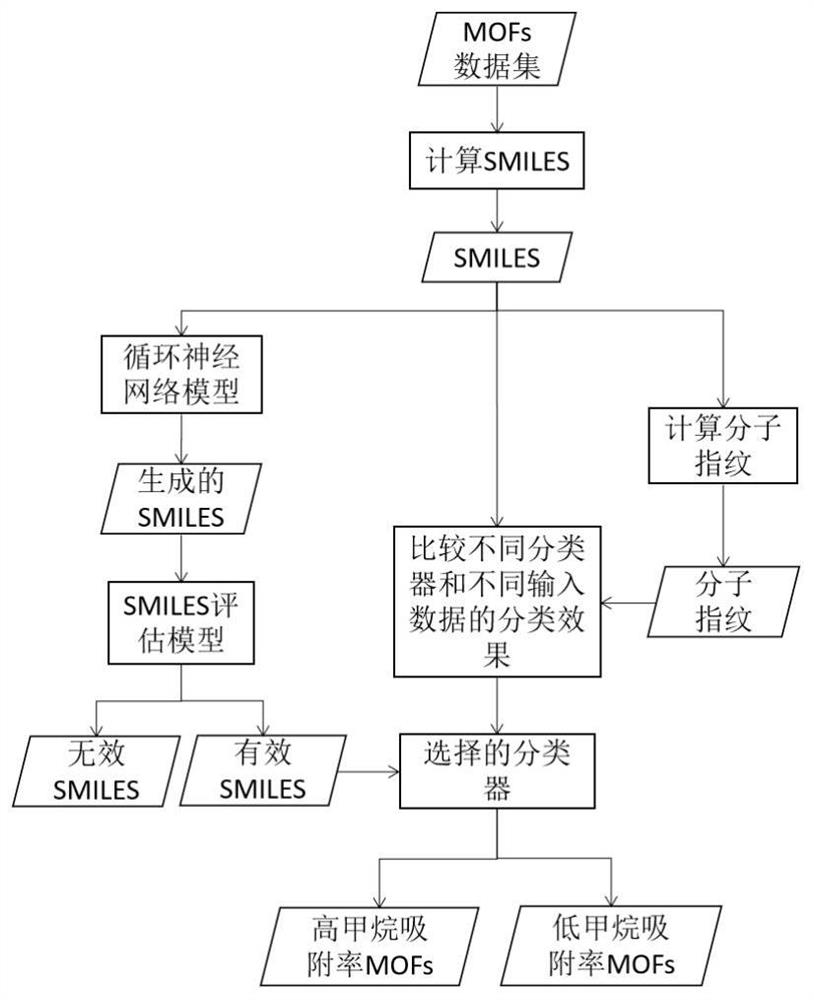
Method for generating and screening MOFs with high methane adsorption rate
PendingCN111755081AShorten the timeSave human effortMathematical modelsKernel methodsData setExperimental laboratory
The invention discloses a method for generating and screening MOFs with a high methane adsorption rate. The method comprises the steps of acquiring simplified molecular linear input specifications ofMOFs in a data set, and calculating molecular fingerprints through SMILES; generating an SMILES representation form of a large number of hypothetical MOFs by using a recurrent neural network model; designing an evaluation model to judge the effectiveness of the generated sample; designing and comparing classification effects of different classifiers and different input data, and screening out theclassifier with the optimal performance; finally, using the trained classifier for predicting the methane adsorption performance of the samples evaluated as effective hypothesis MOFs, screening out the hypothesis MOFs with the high methane adsorption rate, and providing a reference for synthesizing the MOFs with the high methane adsorption rate in a laboratory.
Owner:BEIJING UNIV OF CHEM TECH
Molecular fingerprinting methods to detect and genotype DNA targets through polymerase chain reaction
PendingUS20200123598A1Easy to detectEasily self-collectMicrobiological testing/measurementDisease diagnosisSingle strandGenotyping
A molecular fingerprinting method is disclosed to detect and genotype pathogens DNA targets in a sample through polymerase chain reaction (PCR). The method comprises performing PCR amplification and then a High Resolution Melting (HRM) analysis. The PCR reaction mixture used comprises two or more pairs of amplification primers designed in order to generate amplicons with a different melting temperature each other in order to discriminate, in the HRM analysis, each amplicon by observing the specific melting temperature of each amplicon. The method further comprises monitoring, during the HRM analysis, the change in the signal emission resulting from the temperature-induced denaturation of the double-stranded amplicons into two single-stranded DNAs, due to the release of the intercalating molecule or compound. Discrimination and genotyping of different strains of the same pathogen, different pathogens belonging to separated genus and genetic variations in the sample can be determined through a reader analysing the signal variation; the result of the analysis is obtained through a graphic interface connected to the reader.
Owner:ULISSE BIOMED
SNP locus multicolor fluorescence detection primer and kit used for identifying eucalyptus clones, detection method and applications
ActiveCN110093436AImprove experimental efficiencyEasy to readMicrobiological testing/measurementAgainst vector-borne diseasesFluorescenceGermplasm
The invention discloses a SNP locus multicolor fluorescence detection primer and kit used for identifying eucalyptus clones, a detection method and applications. SNP markers with high polymorphism arescreened in a eucalyptus genome; multicolor fluorescence multiple detection systems of 21 loci are optimized and established; and typing results have the characteristics of being easy in interpretation of banding patterns, high in locus polymorphism and easy in fluorescence distinguishing. The method has the advantages of being high in experimental efficiency, accurate in detection and convenientin operation; important technical support can be provided for rapidly performing germplasm resource genetic evaluation, molecular marker-assisted breeding and construction and identification of clonal molecular fingerprints on eucalyptus in the future; the molecular fingerprints of 58 eucalyptus clones commonly used in China are constructed, identification can be successfully performed on the above clones, so that the workload of eucalyptus clone identification can be greatly reduced; and the method can effectively discriminate counterfeit and disordered clones, so that the rights and interests of breeding cultivation people and forest culture and management planters can be practically guaranteed.
Owner:RES INST OF TROPICAL FORESTRY CHINESE ACAD OF FORESTRY
Method for identifying cytoplasm type of three-line sterile rice by detecting chloroplast DNA (deoxyribose nucleic acid)
InactiveCN102605054AEasy to identifyEfficient identificationMicrobiological testing/measurementBiotechnology3-deoxyribose
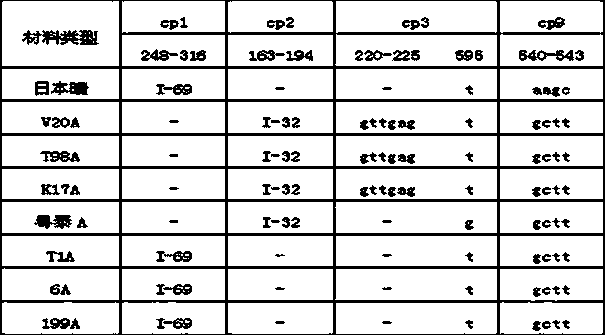
The invention discloses a method for identifying the cytoplasm type of three-line sterile rice by detecting chloroplast DNA (deoxyribose nucleic acid). The method comprises the steps of: taking rice leaves as material, extracting chloroplast DNA in the leaves, carrying out PCR (polymerse chin rection) amplification by using chloroplast primers cp1, cp2, cp3 and cp9, cloning and sequencing the amplified product, and comparing the result with chloroplast DNA molecular fingerprinting of the three-line sterile rice so as to simply and efficiently identify the cytoplasm type of the three-line sterile rice.
Owner:HUNAN NORMAL UNIVERSITY
Genetic fingerprinting and identification method
The present disclosure provides methods for molecular fingerprinting for the characterization and identification of organisms. More specifically, in one aspect the present invention provides a method of identifying an organism in a sample by embedding fingerprint bands from any amplification based fingerprinting method within a DNA sequence so that small differences in size are resolvable. Fingerprint output is provided in a text file format that can then be analyzed by bioinformatics tools.
Owner:QUALICON DIAGNOSTICS LLC
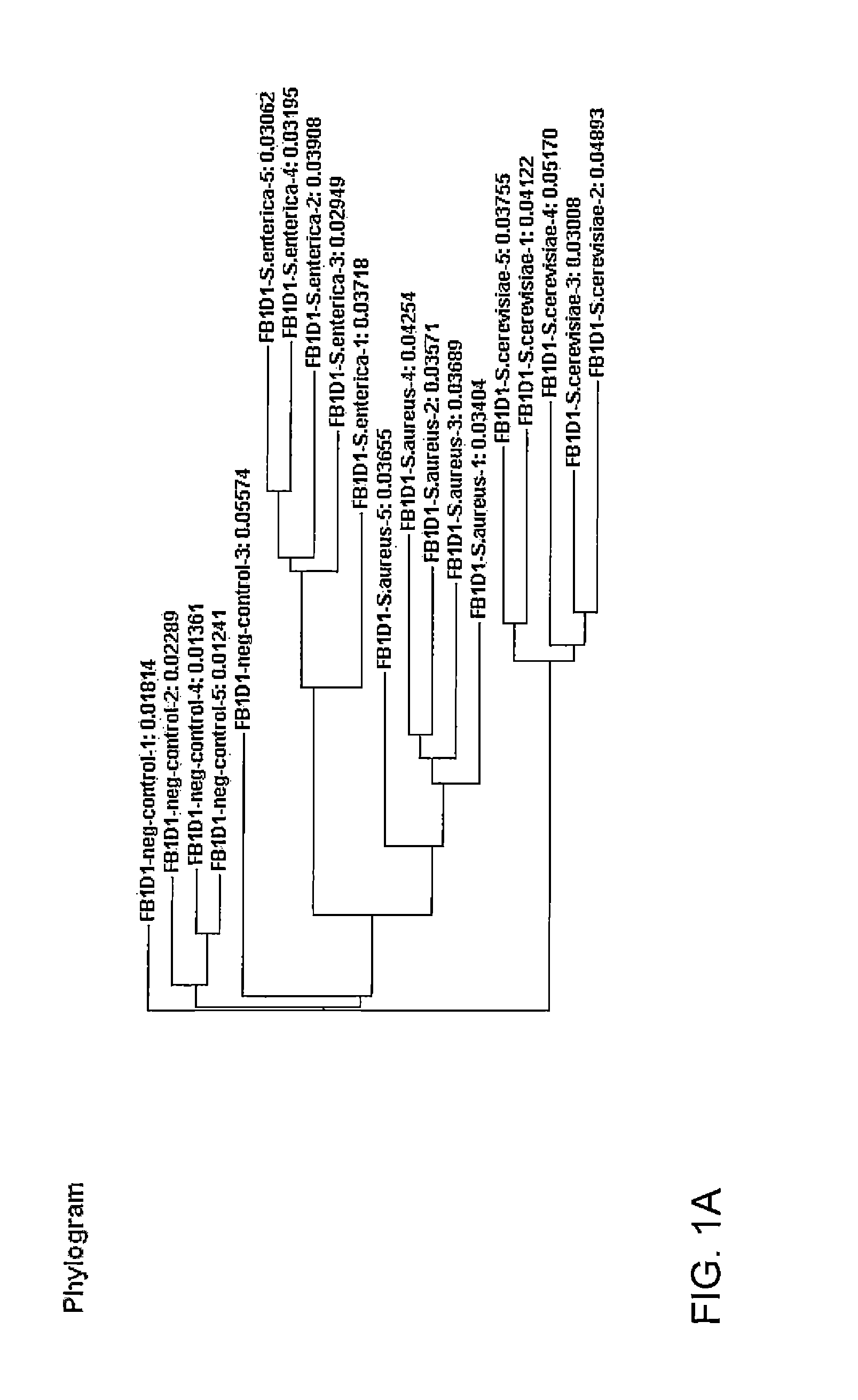
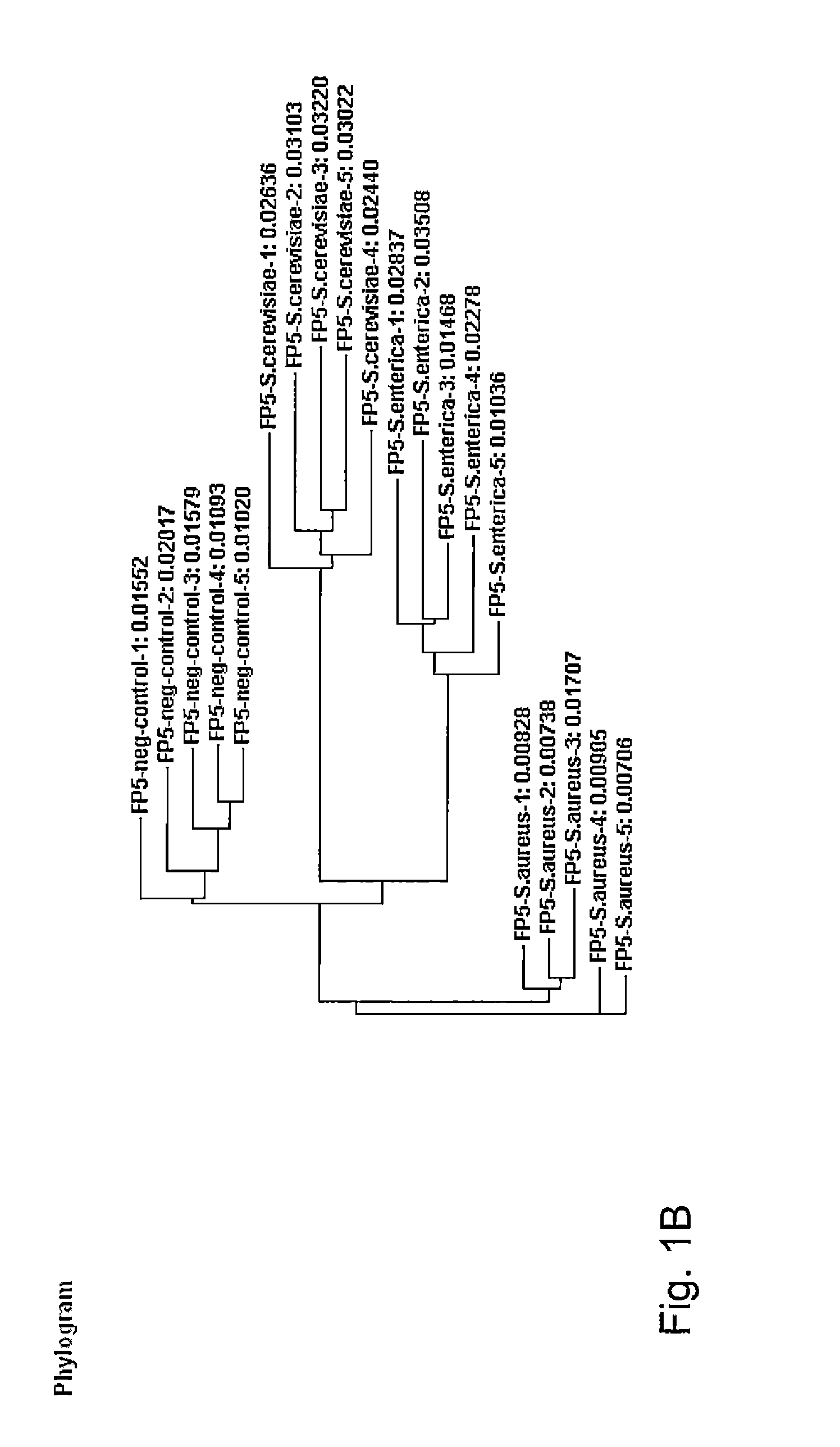
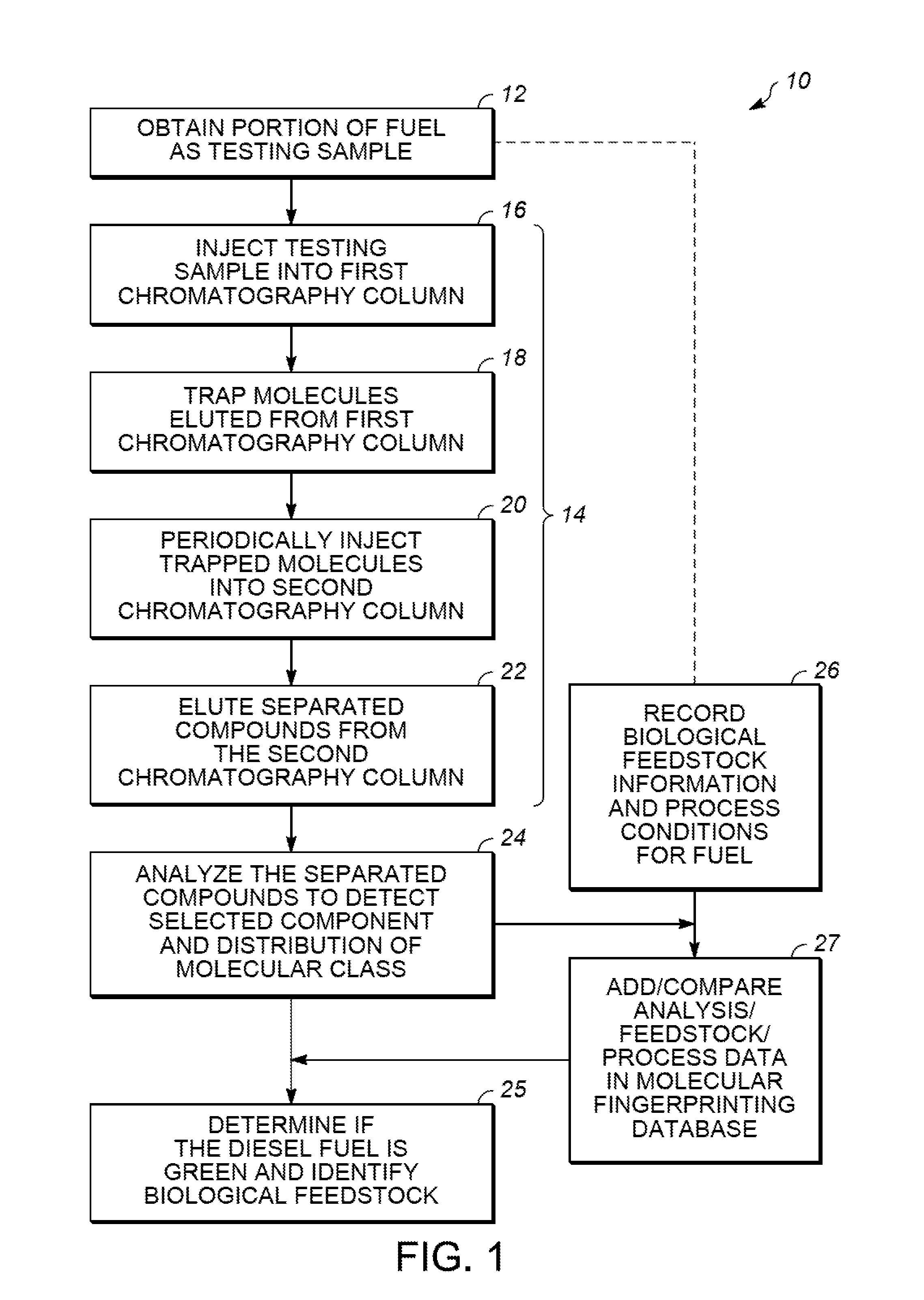
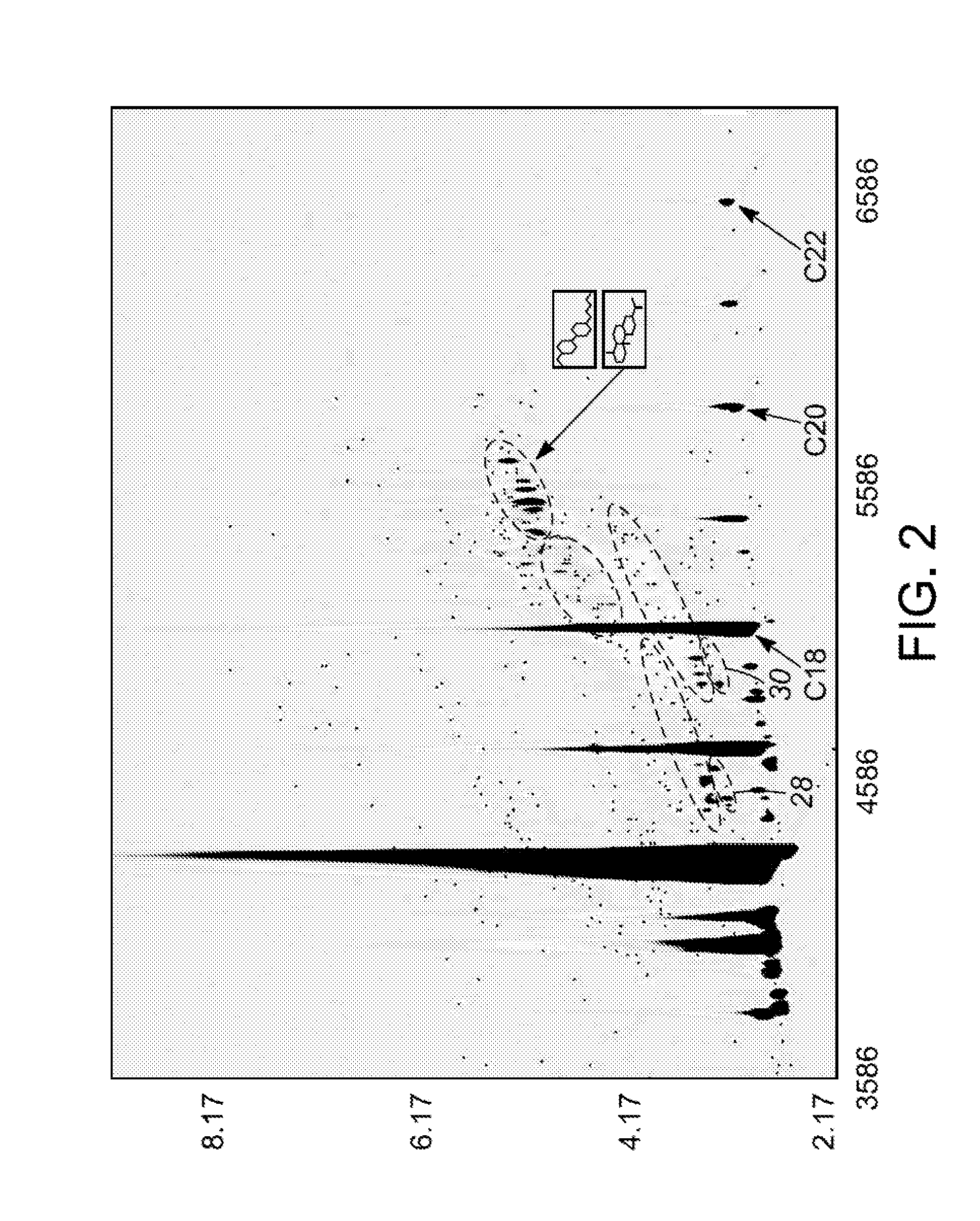
Methods for evaluating green diesel fuel compositions
InactiveUS20130158890A1Component separationSurface/boundary effectMolecular FingerprintingDiesel fuel
Methods for evaluating a diesel fuel are provided. In one embodiment, a method of evaluating a diesel fuel includes obtaining a testing sample from the diesel fuel and analyzing the testing sample to detect an amount of at least one selected component. The method also provides for determining if the diesel fuel is green depending on either detection of at least one selected component in the testing sample or observation of at least one non-normal molecular class distribution in the testing sample. Further, the specific component can indicate the specific biological feedstock used to produce the green diesel based on the molecular fingerprinting database.
Owner:UOP LLC
Method for identifying cytoplasm type of three-line sterile rice by detecting chloroplast DNA (deoxyribose nucleic acid)
InactiveCN102605054BEasy to identifyEfficient identificationMicrobiological testing/measurementBiotechnology3-deoxyribose
The invention discloses a method for identifying the cytoplasm type of three-line sterile rice by detecting chloroplast DNA (deoxyribose nucleic acid). The method comprises the steps of: taking rice leaves as material, extracting chloroplast DNA in the leaves, carrying out PCR (polymerse chin rection) amplification by using chloroplast primers cp1, cp2, cp3 and cp9, cloning and sequencing the amplified product, and comparing the result with chloroplast DNA molecular fingerprinting of the three-line sterile rice so as to simply and efficiently identify the cytoplasm type of the three-line sterile rice.
Owner:HUNAN NORMAL UNIVERSITY
Pharmacokinetic Properties and Toxicity Prediction Method of Drug Molecules Based on Capsule Network
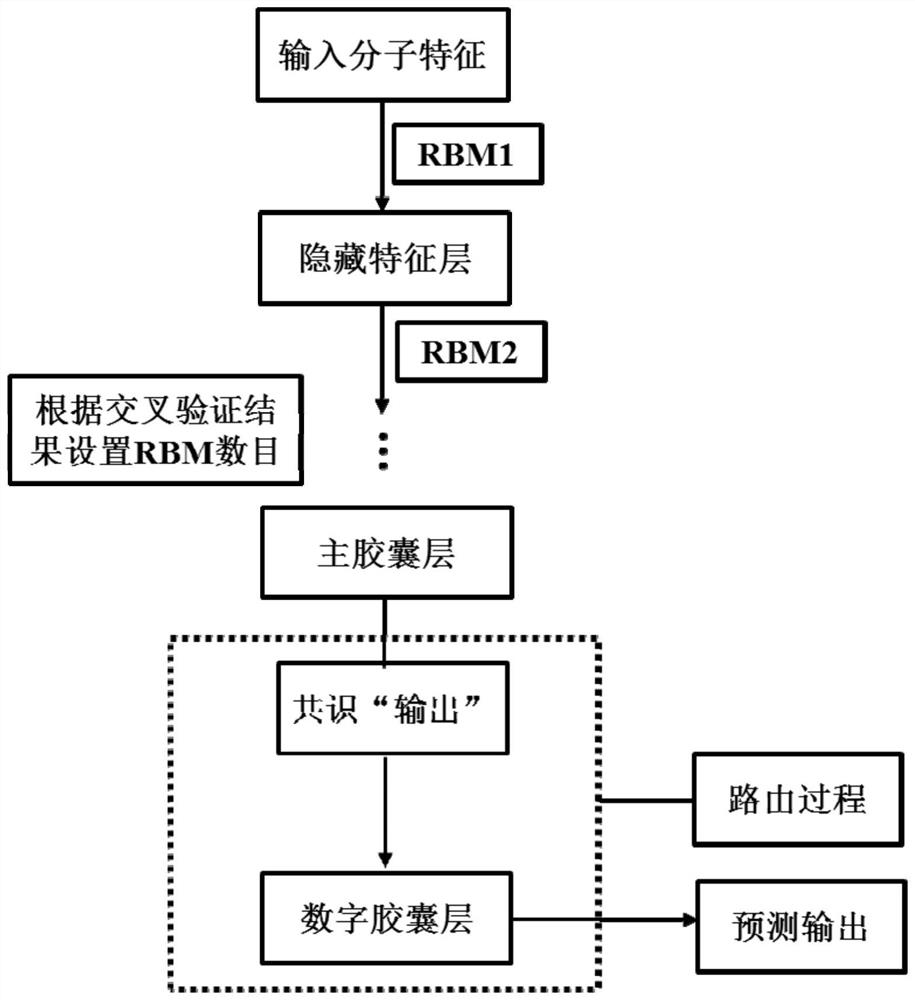
InactiveCN109979541BGood forecastPreserve Hierarchical Positional RelationshipsChemical property predictionComputational theoretical chemistryRestricted Boltzmann machineData set
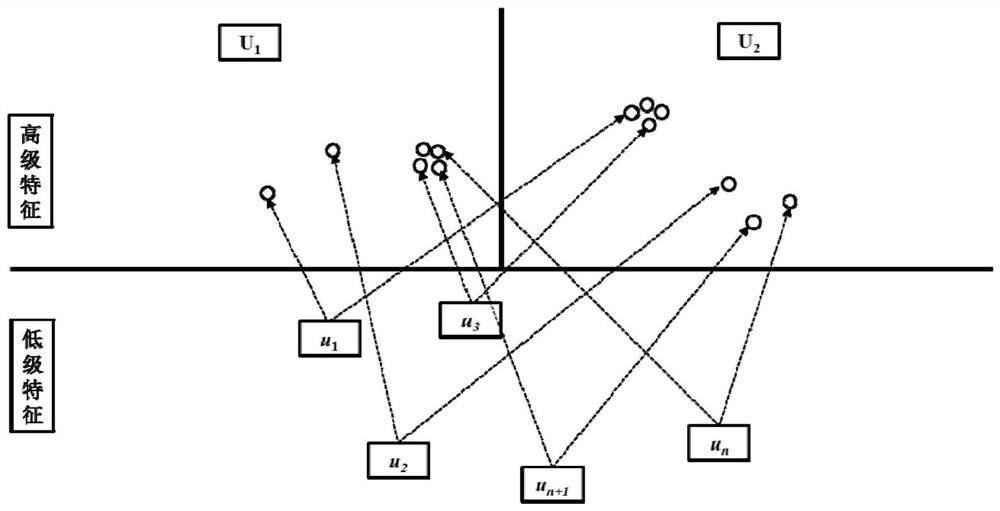
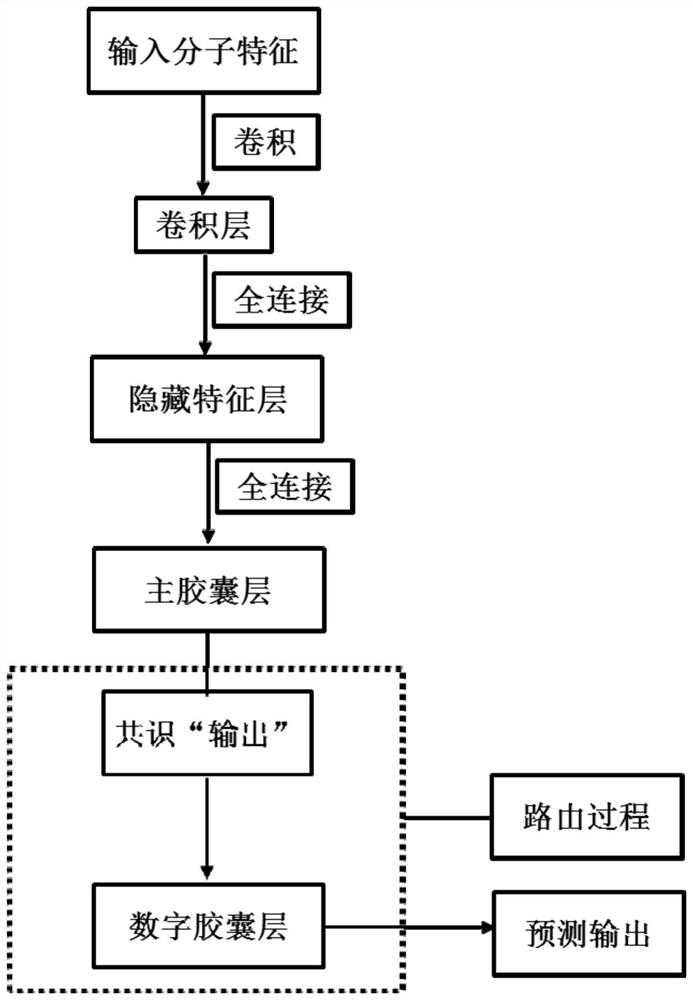
Pharmacokinetic Properties and Toxicity Prediction Method of Drug Molecules Based on Capsule Network. After constructing comprehensive molecular fingerprints and molecular descriptors and preparatory work for establishing models, the low-level feature content of molecules is extracted from the upper-level low-level features through convolution or restricted Boltzmann machine operations, and then the capsule network method is used in the next step. The high-level features of molecules are abstracted from a layer of high-level features, and the relationship between high-level features and active labels is fitted by a dynamic routing algorithm to predict the pharmacokinetic properties and toxicity classification of unknown small molecules. The present invention does not need to collect large-scale data sets for training, and realizes automatic dimensionality reduction through end-to-end optimization of the input, updates the coupling coefficient through iterative dynamic routing process, and dynamic routing transfers all the features of the upper layer capsule to any one of the lower layer In the capsule, the hierarchical position relationship between the low-level features and high-level features is greatly preserved. Better predictive performance than traditional machine learning methods.
Owner:SICHUAN UNIV
SSR multiple detection primer, kit and method for pithecellobium clypearia variety identification
PendingCN114277172AImprove economyReduce experiment costsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMedicinal herbs
The invention discloses SSR multiple detection primers, a kit and a method for pithecellobium clypearia variety identification. Compared with independent PCR detection of a single marker, the method has the advantages of good economical efficiency, reagent saving, time saving, high efficiency and the like, and a simple and reliable method is provided for population diversity analysis of pithecellobium clypearia and sibling species, genetic evaluation of germplasm resources, authenticity identification of varieties and medicinal materials and the like; the method is used for construction of SSR molecular fingerprints of pithecellobium clypearia varieties (cutting clones) and identification of to-be-detected varieties, has the advantages of being accurate, reliable, effective and rapid, can effectively discriminate counterfeit and disordered varieties and new varieties, and provides important technical support for guaranteeing rights and interests of improved variety cultivators and forest planters.
Owner:RES INST OF TROPICAL FORESTRY CHINESE ACAD OF FORESTRY
Method for extracting the total microbial genome in vinegar fermentation process
ActiveCN103409410BEasy to settleIntegrity guaranteedMicrobiological testing/measurementDNA preparationBiotechnologyGenomics
The invention relates to a general method for extracting the total microbial genome form traditional vinegar fermentation process samples, and belongs to the technical filed of molecular biology. A technical issue to be solved by the invention is to provide a method for extracting the total microbial genome from traditional vinegar samples in different states comprising solid state fermentation, semi-solid state fermentation and liquid state fermentation. The method comprises steps of pretreating the vinegar fermentation samples, extracting microbial genome DNA, removing impurities, rinsing and concentrating. The method for extracting relates to in-situ extraction. The sample amount needed is less than 5 g. Addition of preparations such as protease, cellulase, helicase, etc. is not needed. The genome extracted has high purity and high quality, and can be directly used for subsequent molecular fingerprint analysis, metagenomics analysis, PCR (polymerase chain reaction) amplification of targeted genes and clone library construction.
Owner:TIANJIN UNIV OF SCI & TECH
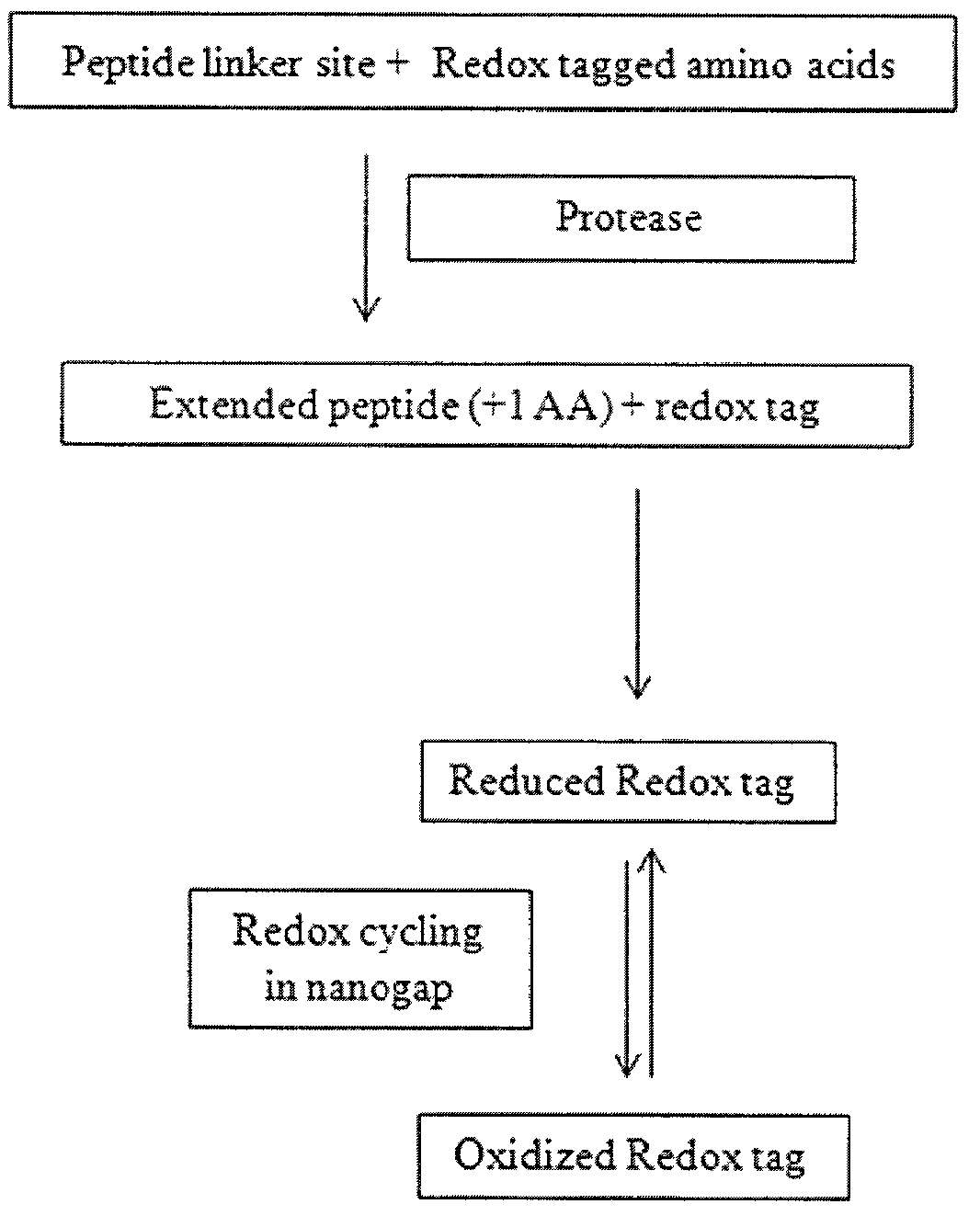
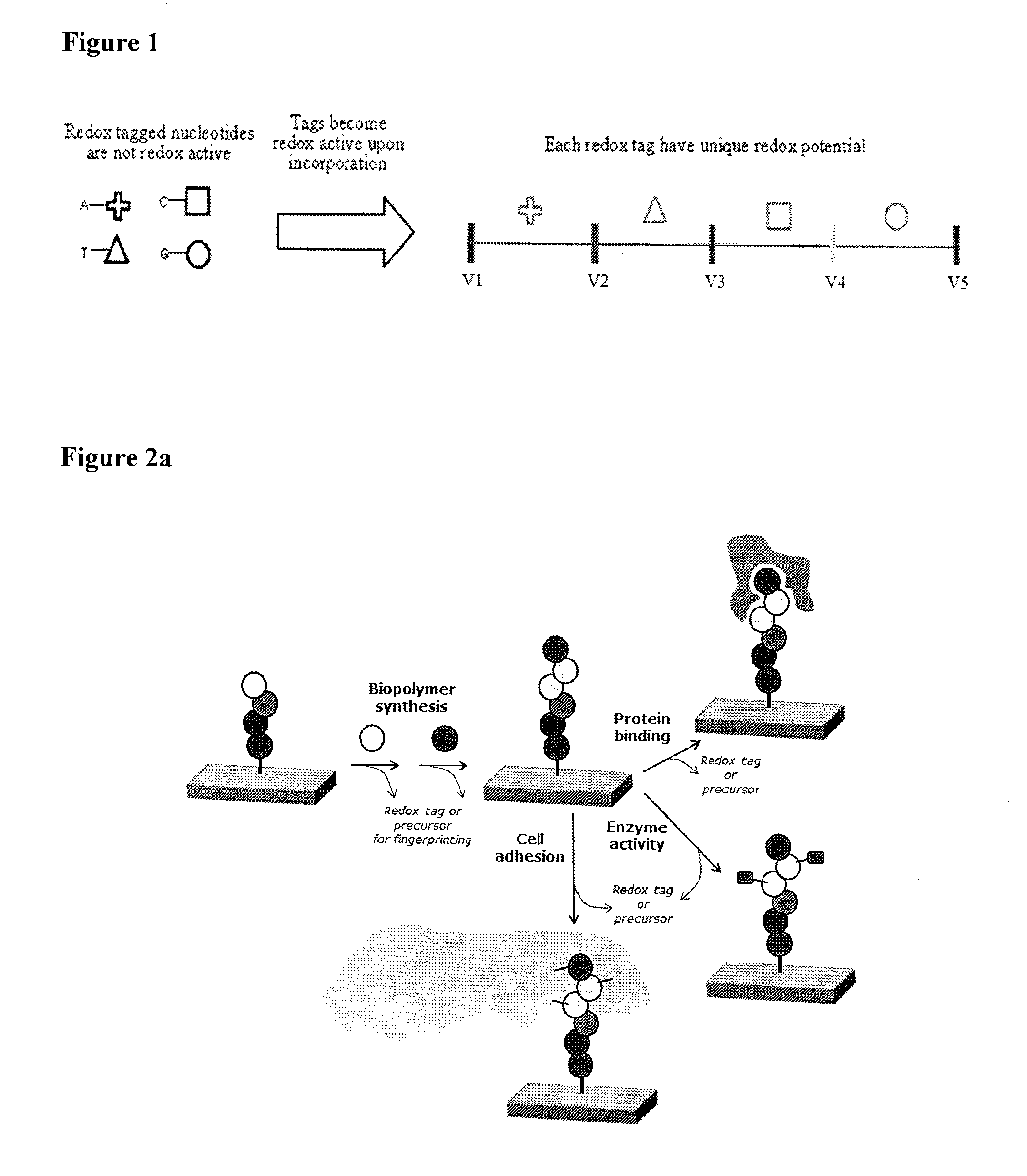
High throughput biochemical detection using single molecule fingerprinting arrays
InactiveUS9551682B2Sequential/parallel process reactionsMaterial analysis by electric/magnetic meansBio moleculesChemical reaction
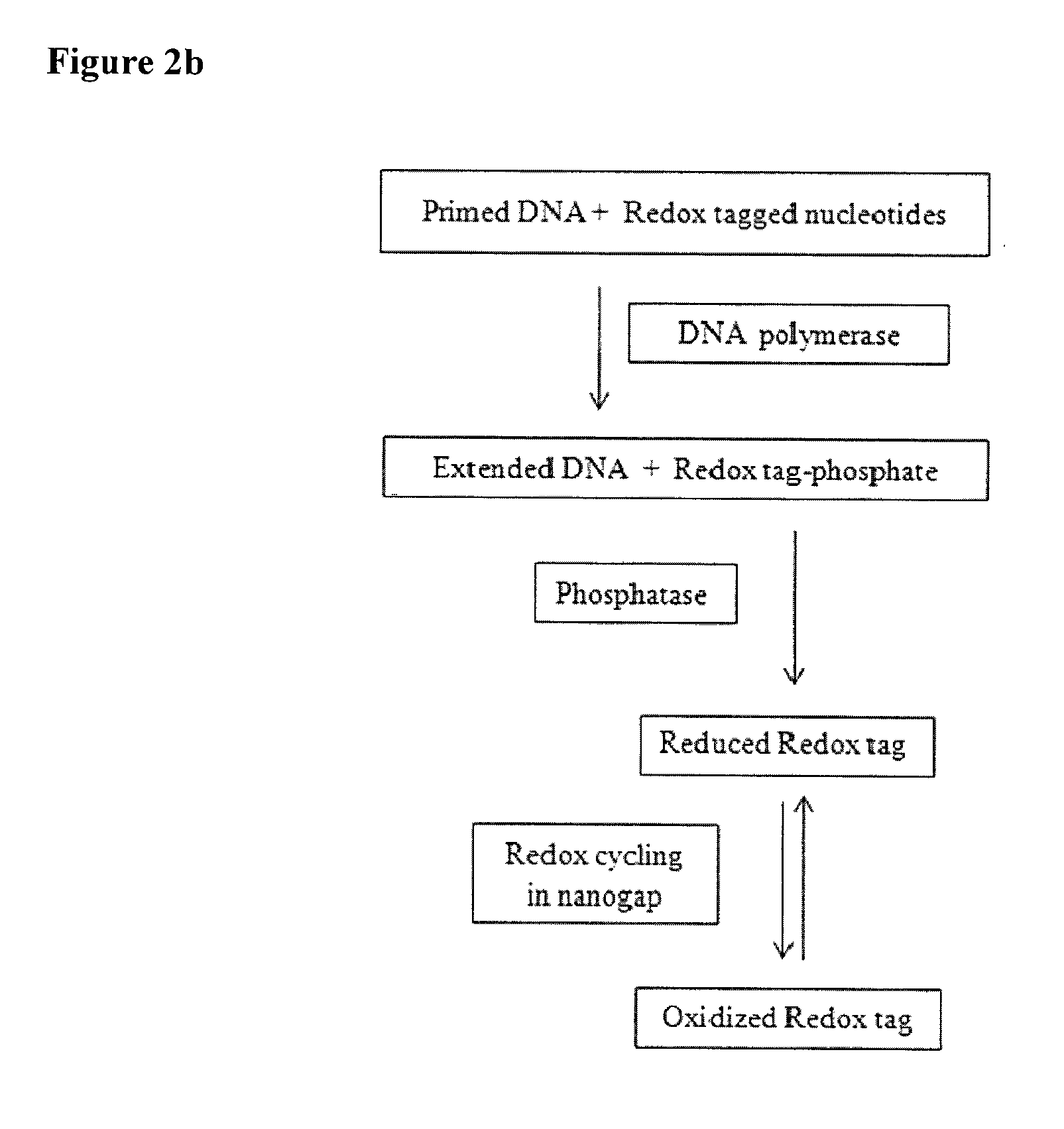
Various embodiments provide devices, methods, and systems for high throughput biomolecule detection using transducer arrays. In one embodiment, a transducer array made up of transducer elements may be used to detect byproducts from chemical reactions that involve redox genic tags. Each transducer element may include at least a reaction chamber and a fingerprinting region, configured to flow a fluid from the reaction chamber through the fingerprinting region. The reaction chamber can include a molecule attachment region and the fingerprinting region can include at least one set of electrodes separated by a nanogap for conducting redox cycling reactions. In embodiments, by flowing the chamber content obtained from a reaction of a latent redox tagged probe molecule, a catalyst, and a target molecule in the reaction chamber through the fingerprinting region, the redox cycling reactions can be detected to identify redox-tagged biomolecules.
Owner:INTEL CORP
Construction method of ophiopogon japonicus AFLP molecular fingerprint spectrum library and application of construction method
PendingCN112080496AQuality improvementHigh resolutionMicrobiological testing/measurementDNA/RNA fragmentationOphiopogon japonicusGermplasm
The invention discloses a primer pair combination, and further discloses application of the primer pair combination in ophiopogon japonicus germplasm identification or variety identification or construction of an AFLP molecular fingerprint spectrum library. The invention further discloses a construction method and application of the ophiopogon japonicus AFLP molecular fingerprint spectrum library.The selective amplification primers and primer combinations capable of effectively identifying different germplasms of ophiopogon japonicus and the high-quality DNA fingerprint spectrum are successfully screened through repeated unloosened tests, so that the method has the advantages of multiple amplification sites, good band type quality, high resolution, proper band density, uniform distribution and easiness in accurately detecting the subtle difference of samples, so that a technical foundation is laid for constructing the ophiopogon japonicus AFLP molecular fingerprint spectrum library; and 30 ophiopogon japonicus with different germplasms can be successfully and effectively identified by nine pairs of primer combinations and the like through a provided reagent method, and a scientific basis is provided for ophiopogon japonicus germplasm identification, new variety registration, registration, property right protection and the like in the future.
Owner:JIANGSU POLYTECHNIC COLLEGE OF AGRI & FORESTRY
Genetic Fingerprinting And Identification Method
InactiveUS20110045478A1Size differenceSugar derivativesMicrobiological testing/measurementBiological bodyA-DNA
The present disclosure provides methods for molecular fingerprinting for the characterization and identification of organisms. More specifically, in one aspect the present invention provides a method of identifying an organism in a sample comprising: (a) providing a sample comprising said organism, said organism comprising at least one nucleic acid; (b) combining said sample or the at least one nucleic acid therefrom with an amplification mix comprising at least one labeled oligonucleotide primer; (c) generating at least one labeled amplification product from the at least one nucleic acid of said organism using a nucleotide amplification technique employing said at least one labeled oligonucleotide primer; (d) combining said at least one labeled amplification product with products of a DNA sequencing reaction to create a separation mix; and (e) separating said separation mix on the basis of oligonucleotide length in a fluorescent DNA sequencing instrument to generate a sequence embedded fingerprint pattern for said organism.
Owner:QUALICON DIAGNOSTICS LLC
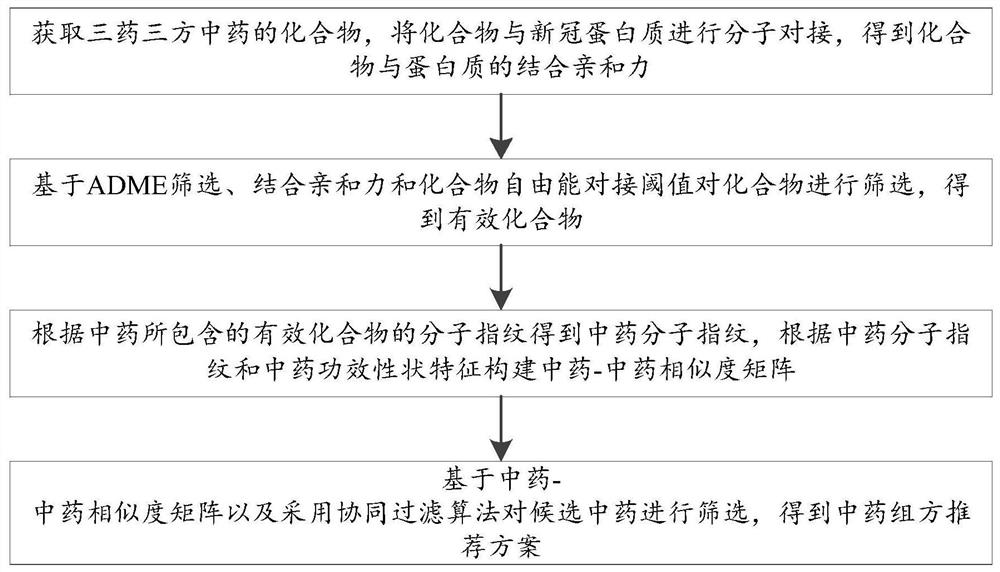
Method and system for recommending traditional Chinese medicine prescriptions for novel coronavirus based on collaborative filtering
ActiveCN112667922BImprove bindingLower free energyChemical property predictionDigital data information retrievalEfficacyTraditional medicine
The invention discloses a novel coronavirus traditional Chinese medicine prescription recommendation method and system based on collaborative filtering, including: obtaining the compound of three medicines and three traditional Chinese medicines, molecularly docking the compound with the new coronavirus protein, and obtaining the binding affinity between the compound and the protein; screening based on ADME , Combining affinity and compound free energy docking threshold to screen compounds to obtain effective compounds; obtain molecular fingerprints of traditional Chinese medicines according to the molecular fingerprints of effective compounds contained in traditional Chinese medicines, and construct traditional Chinese medicine-Chinese medicine similarity matrix according to molecular fingerprints of traditional Chinese medicines and efficacy traits of traditional Chinese medicines; Based on the traditional Chinese medicine-Chinese medicine similarity matrix and the collaborative filtering algorithm, the candidate Chinese medicines were screened, and the recommended prescriptions of Chinese medicines were obtained. Molecular docking of the three-drug and three-prescription Chinese medicine compound and the new crown protein was carried out to measure the binding ability of the traditional Chinese medicine and the traditional Chinese medicine prescription and the new crown protein. At the same time, the recommendation of the traditional Chinese medicine prescription was made based on the collaborative filtering algorithm, and the effectiveness of the Chinese medicine prescription for the treatment of the new crown virus was verified.
Owner:SHANDONG UNIV
A method for rapid assessment of adverse drug reactions based on molecular fingerprints
ActiveCN105787261BIncrease success rateElucidate molecular mechanismsChemical property predictionMolecular designNetwork modelADR - Adverse drug reaction
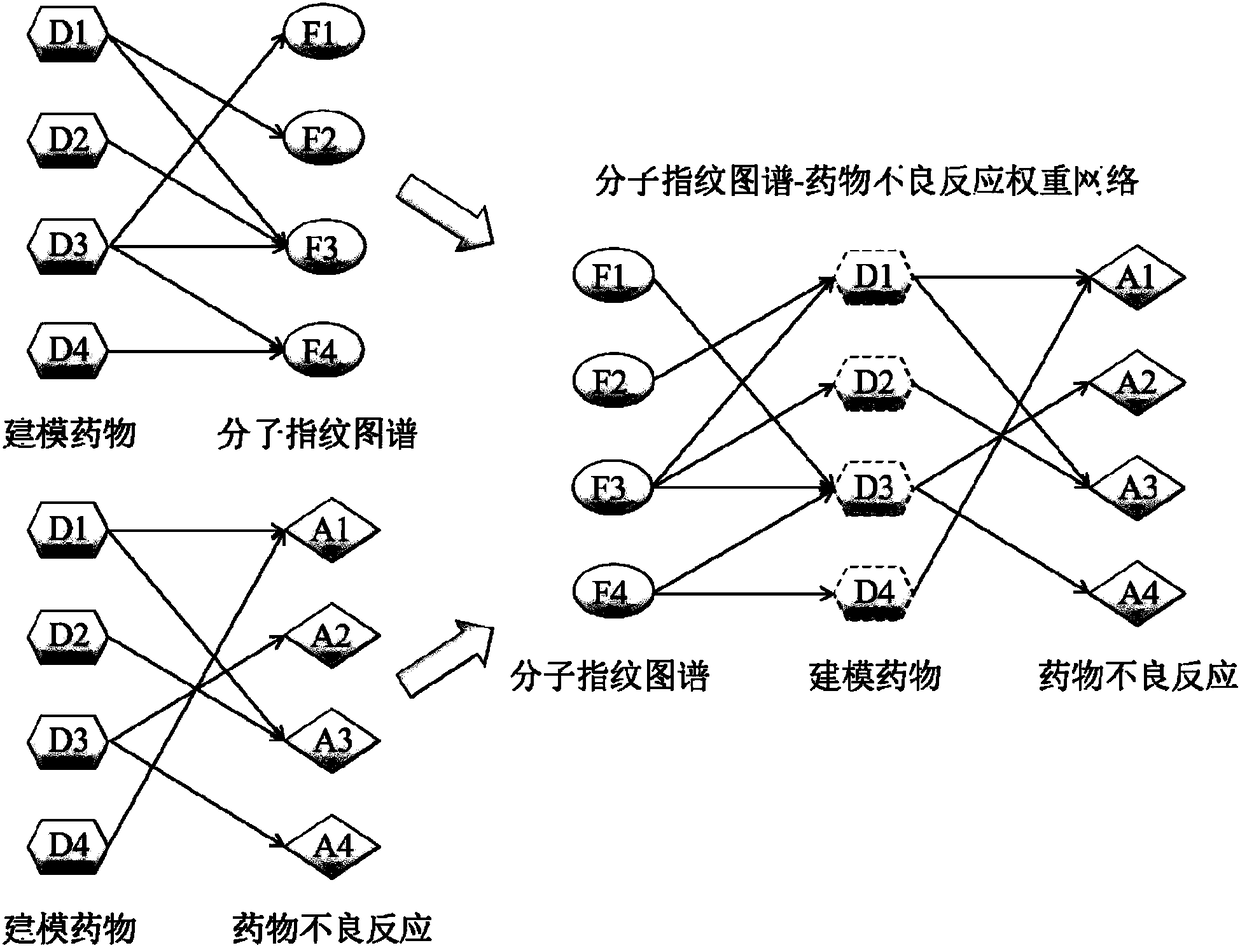
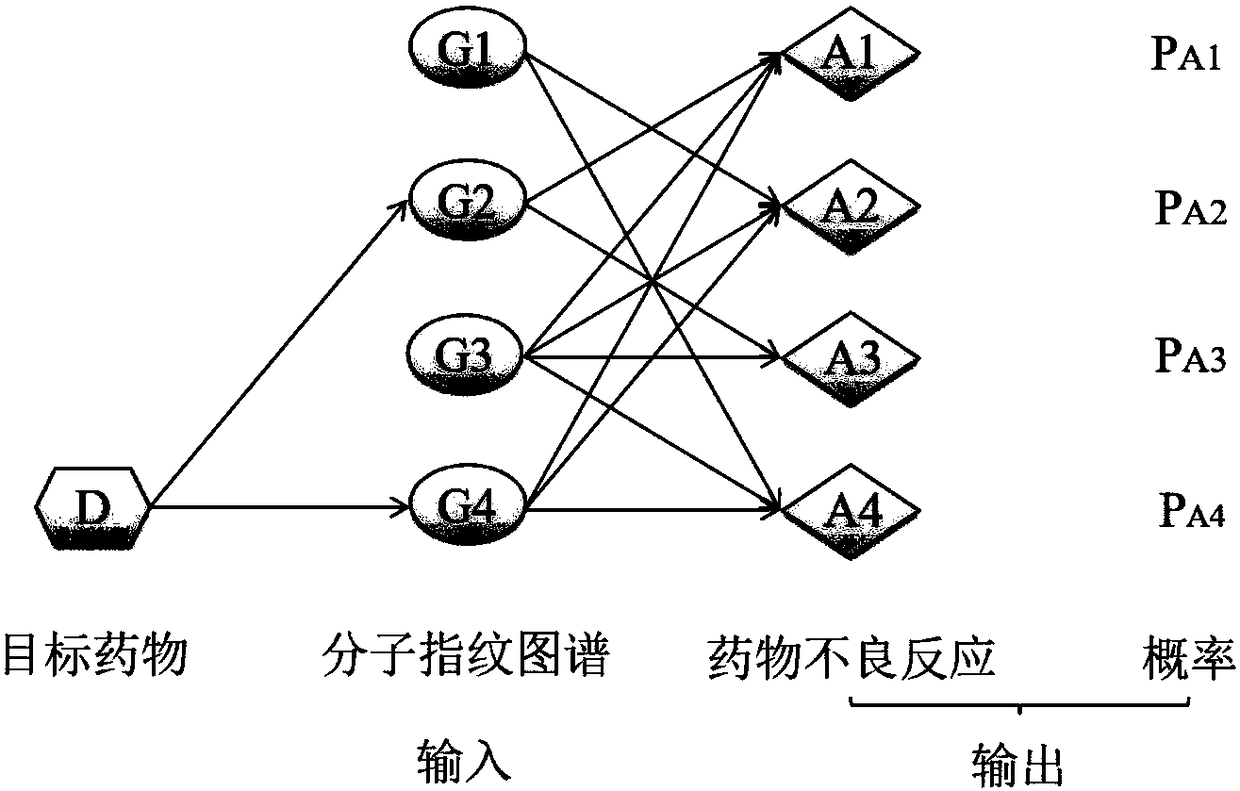
The invention relates to the technical field of computers, in particular to a method for rapidly assessing adverse drug reactions based on a molecule fingerprint spectrum.The method includes the steps that 1, a drug-adverse drug reaction relationship network is built; 2, a drug-molecule fingerprint spectrum corresponding relationship network is built; 3, a drug-molecule fingerprint spectrum-adverse drug reaction network model is built; 4, the probability that the adverse drug reactions A<m> are possibly caused by a target drug D is calculated; 5, the probability P (A<m>|F<multi>) that the adverse drug reactions A<m> are sensed by the fingerprint-spectrum set F<multi> of the target drug D is calculated; 6, the adverse drug reactions with P (A<m>|F<multi>) higher than 1% are screened.The method can serve as an economical and effective method for assessing the adverse drug reactions in a rapid, high-throughput and quantization mode in the early drug research-and-development stage.
Owner:XIAMEN UNIV
Method and device for extracting molecular fingerprints and calculating relevancy based on molecular fingerprints
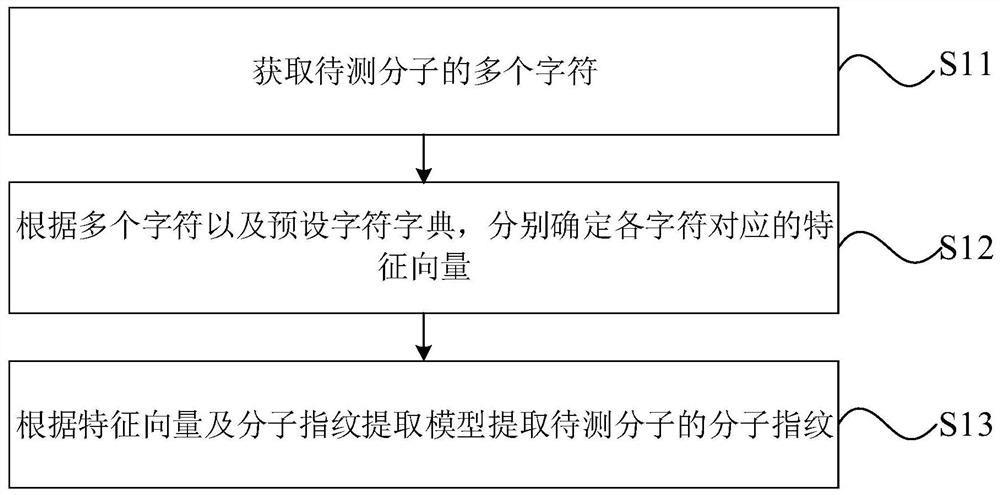
PendingCN112201314AShorten the timeAccurate evaluationMolecular designFeature vectorVirtual screening
The invention discloses a molecular fingerprint extraction method and device and a relevancy calculation method and device based on the molecular fingerprint extraction method, and the molecular fingerprint extraction method comprises the steps: obtaining a plurality of characters of a to-be-detected molecule; according to the plurality of characters and a preset character dictionary, respectivelydetermining a feature vector corresponding to each character; and extracting the molecular fingerprint of the to-be-detected molecule according to the feature vector and the molecular fingerprint extraction model. By implementing the method and the device, the problem that the potential activity aspect of the molecule is not related even if the structure is relatively similar due to the fact thatthe molecular fingerprint determined on the basis of artificially designed molecular characteristics cannot describe the overall structure of the molecule in the related technology is solved, and thekey characteristic information of the molecule is obtained, and the relatively accurate molecular activity relevancy information is obtained, so the molecular similarity can be accurately evaluated,so ligand-based virtual screening can be more accurate and efficient, and the time required by virtual screening is effectively shortened.
Owner:北京望石智慧科技有限公司
Preparation method of one-dimensional single crystal germanium-based graphene plasmon nanostructure
ActiveCN113981533APrevent oxidationAvoid defectsPolycrystalline material growthVacuum evaporation coatingNanowireSingle crystal
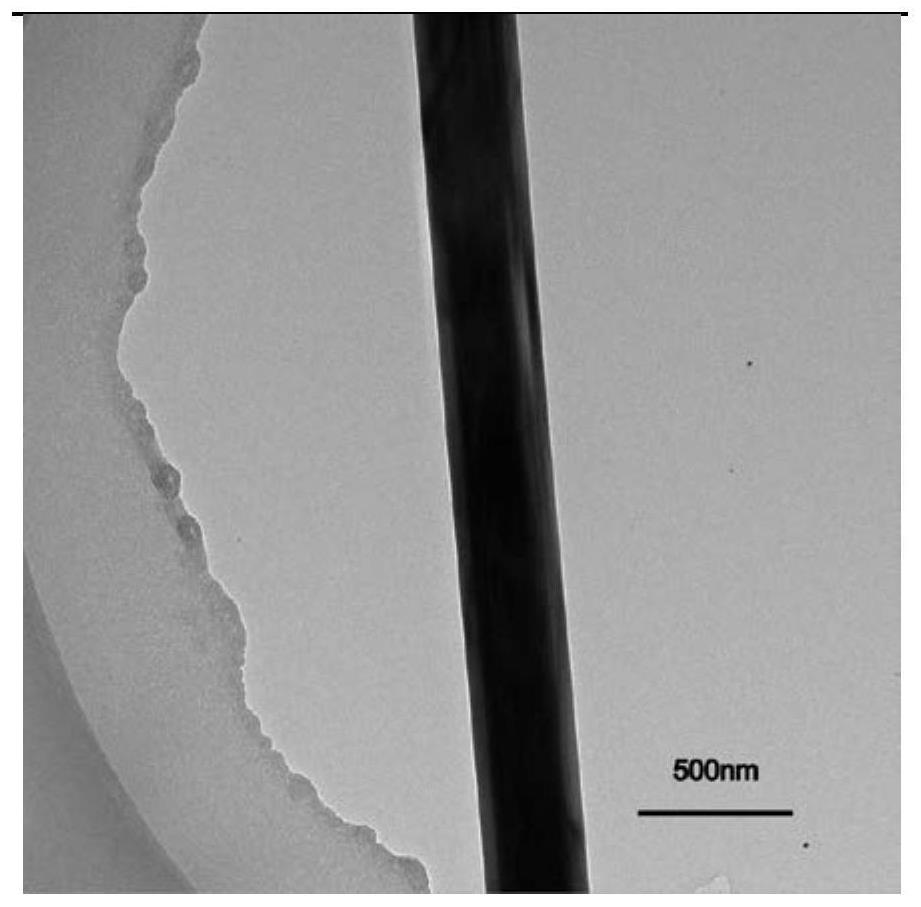
The invention provides a preparation method of a one-dimensional single crystal germanium-based graphene plasmon nanostructure. The preparation method comprises the following steps: (1) sputtering a gold nano film on a substrate by using a magnetron sputtering method; (2) growing a germanium-doped single crystal nanowire on the substrate obtained in the step (1) by adopting chemical vapor deposition; and (3) directly growing high-quality graphene on the surface of the germanium-doped single crystal nanowire obtained in the step (2) by adopting chemical vapor deposition to obtain the one-dimensional germanium-based graphene surface plasmon nanostructure. According to the invention, graphene directly grows on the surface of a germanium single crystal nanowire through a chemical vapor deposition method, so that the traditional graphene film transfer process is avoided, the influence of the transfer process on the carrier mobility of graphene is reduced to the maximum extent, and coupling of mid-infrared surface plasmons between germanium and graphene is enhanced; and the material provided by the invention has a strong and adjustable surface plasmon effect, and is expected to be widely applied to the fields of molecular fingerprint detection, intermediate infrared sensing, photon chips and the like.
Owner:HENAN UNIVERSITY
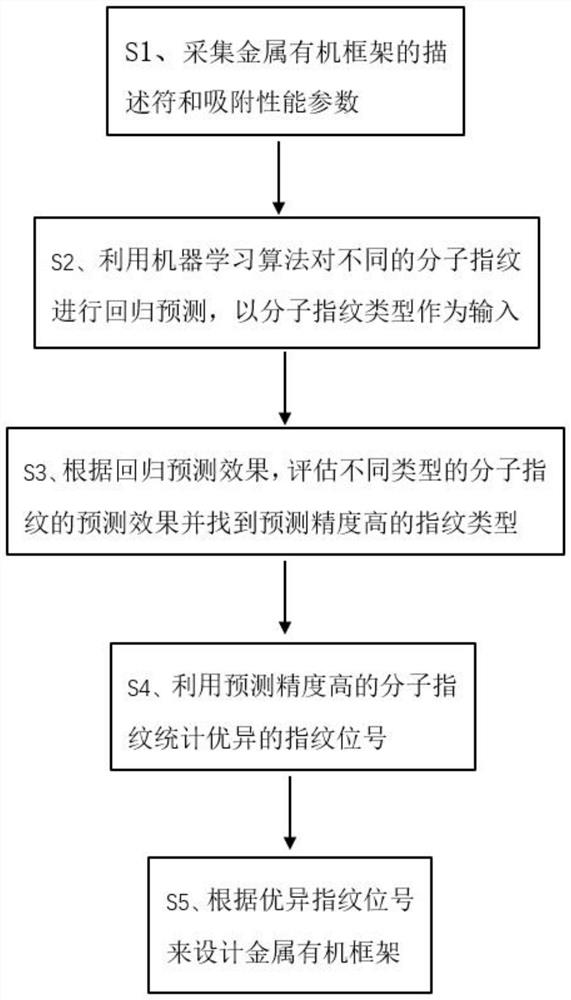
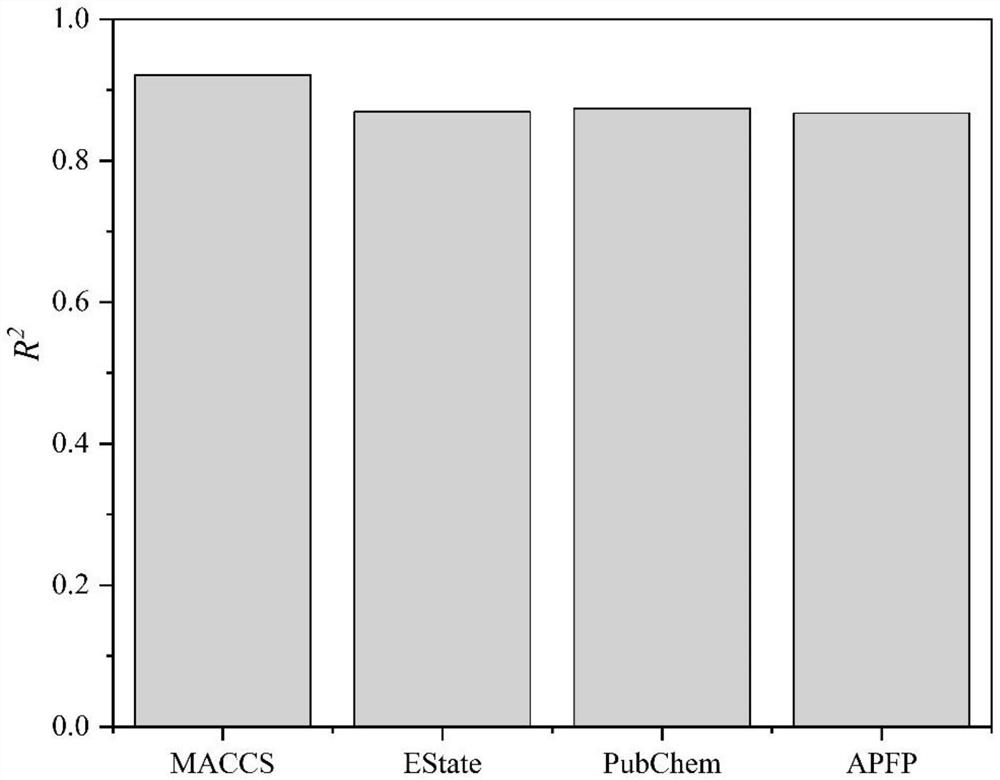
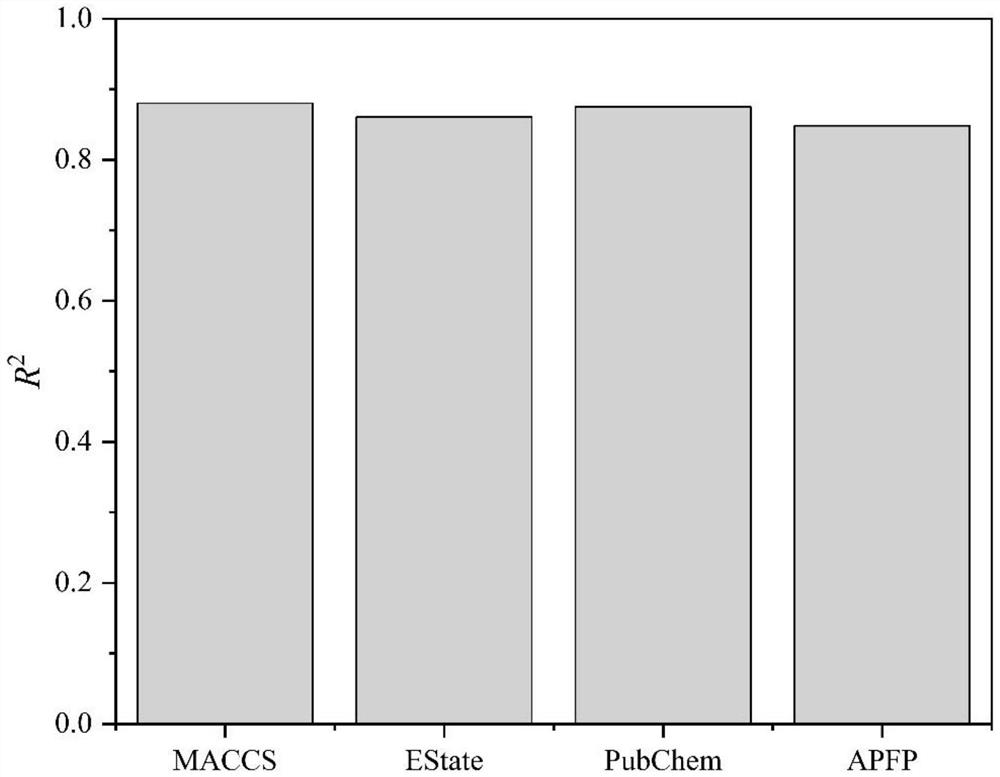
Method for designing metal organic framework based on molecular fingerprints
PendingCN114783534AEfficient designImprove screening efficiencyMolecular designKernel methodsMetal-organic frameworkPubChem
The invention relates to the field of computational chemistry and nano composite catalytic materials, and discloses a method for designing a metal organic framework based on molecular fingerprints, which comprises the following steps of: 1, acquiring descriptors and adsorption performance parameters of the metal organic framework; a second step of performing regression prediction on different molecular fingerprints by utilizing a machine learning algorithm based on the descriptors and the adsorption performance parameters, and taking molecular fingerprint types as input, the molecular fingerprint types including APFP, Estate, MACCS and PubChem; 3, according to a regression prediction effect, evaluating prediction effects of different types of molecular fingerprints, finding out a fingerprint type with high prediction precision, and obtaining a molecular fingerprint with high prediction precision; 4, counting excellent fingerprint bit numbers by using the molecular fingerprints with high prediction precision; and 5, designing the metal organic framework according to the excellent fingerprint bit number. According to the method, the important fingerprint bit number can be accurately and quickly found, and the design of the metal organic framework is guided.
Owner:GUANGZHOU UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com