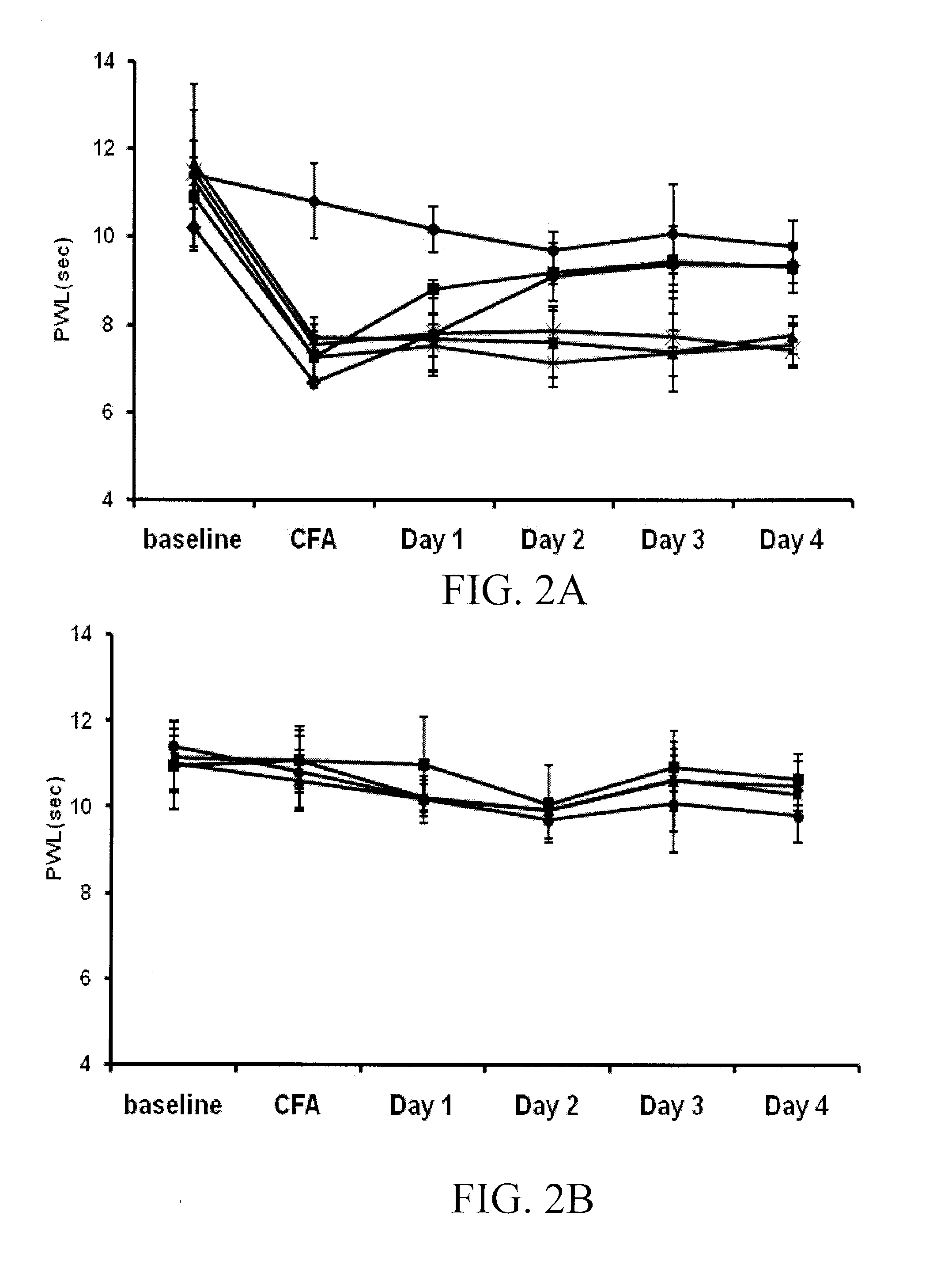
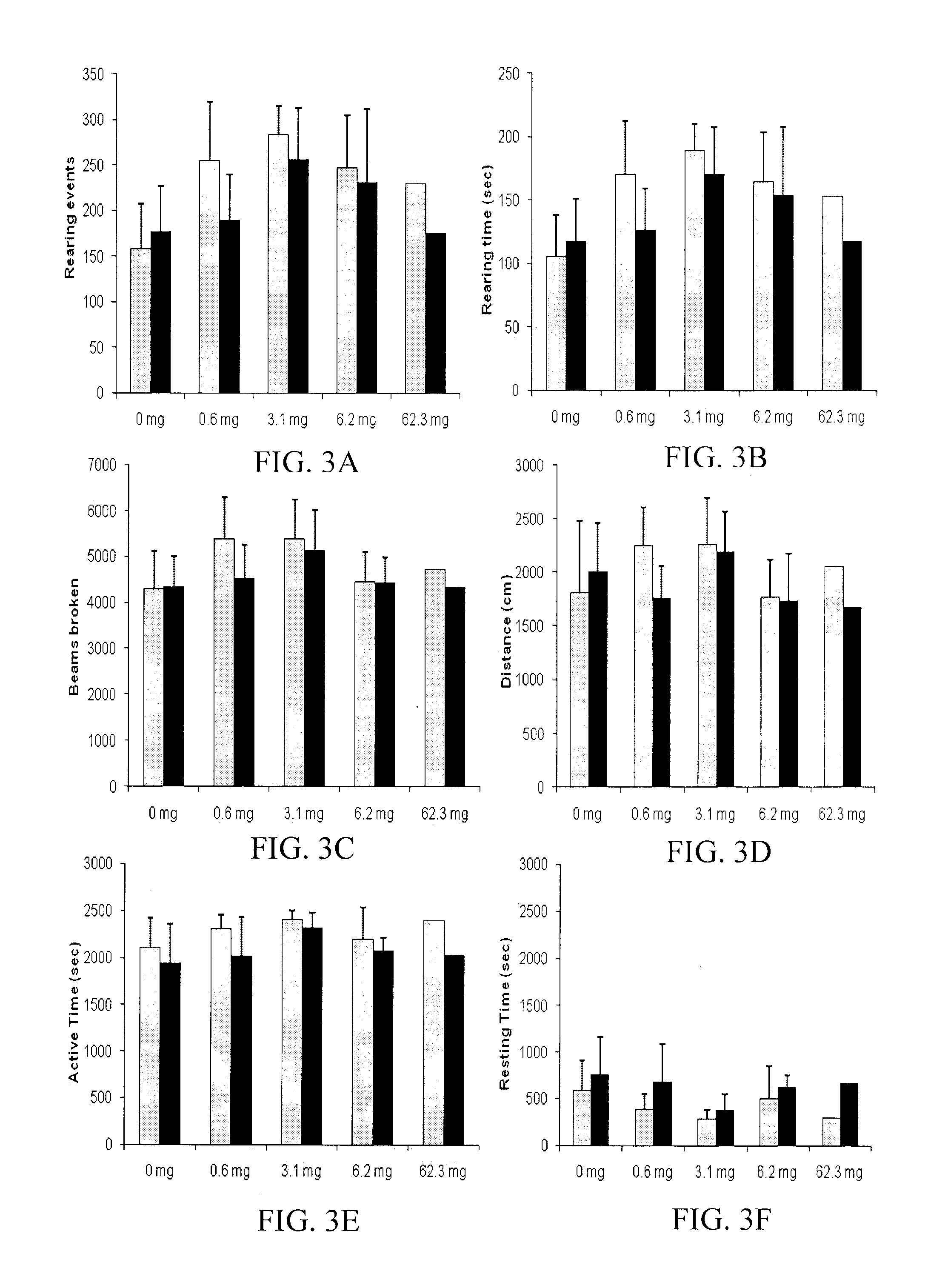
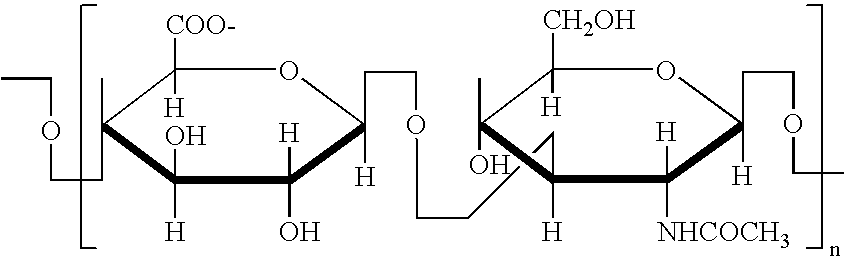
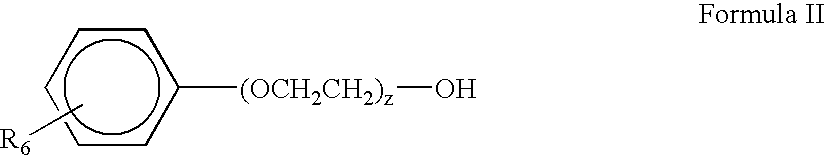Patents
Literature
43 results about "Arthritic joint" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The main symptoms of arthritis are joint pain and stiffness, which typically worsen with age. The most common types of arthritis are osteoarthritis and rheumatoid arthritis. Osteoarthritis causes cartilage — the hard, slippery tissue that covers the ends of bones where they form a joint — to break down.
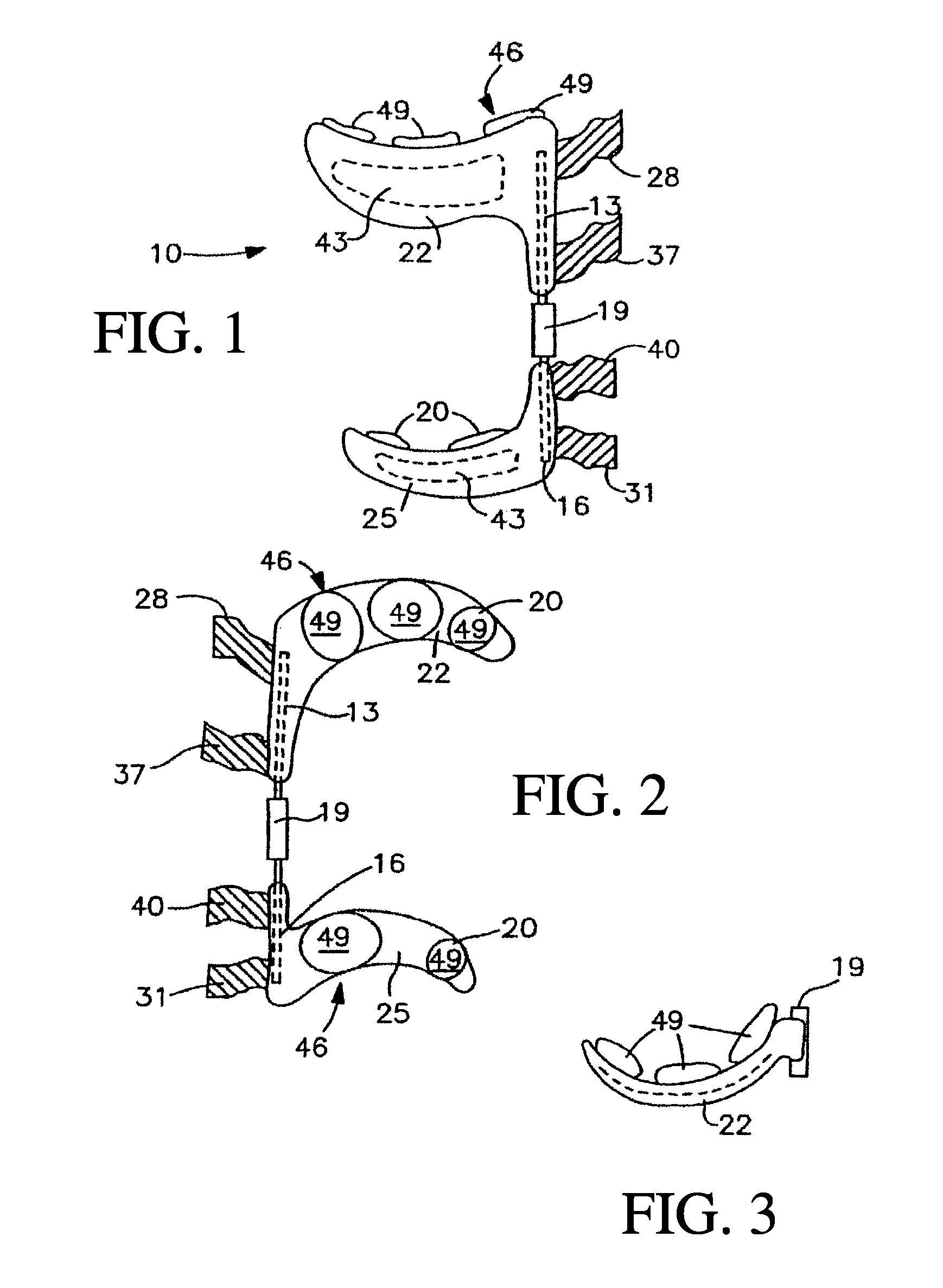
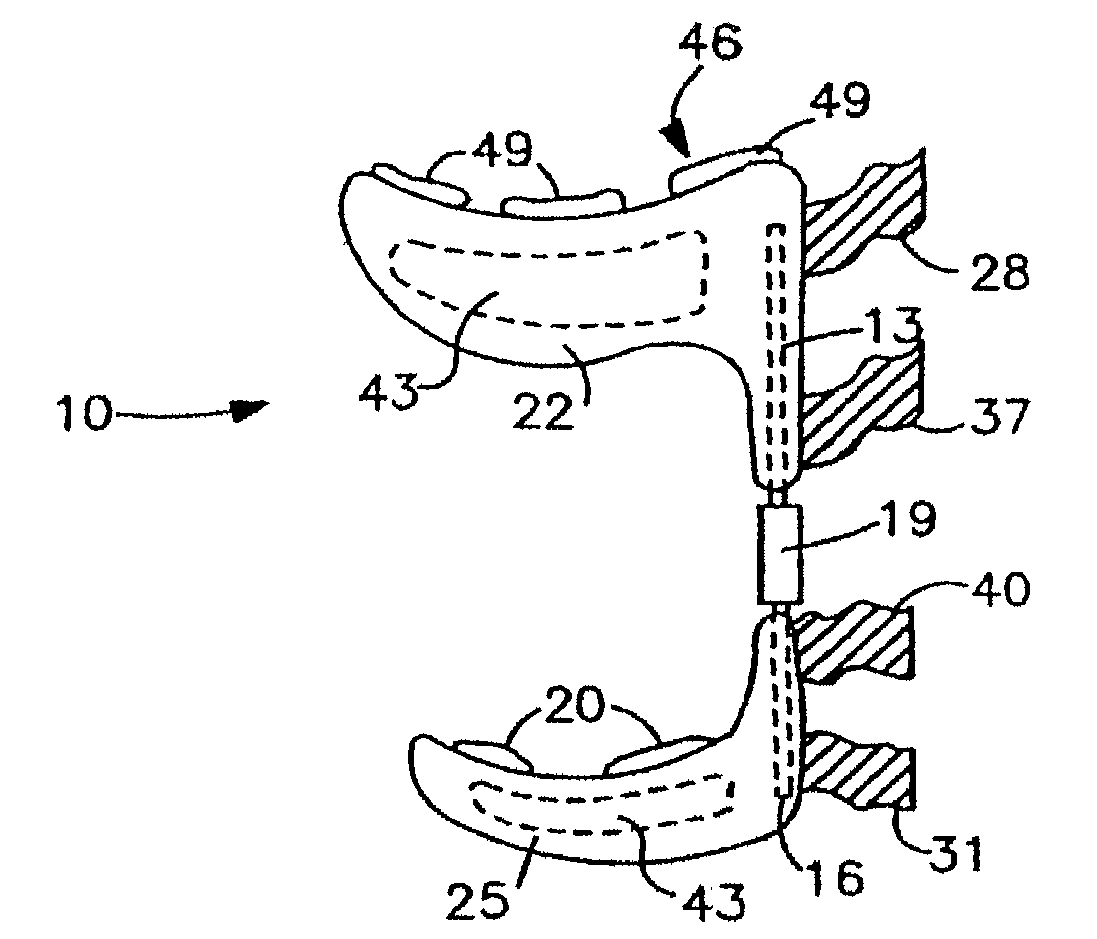
Joint treating method
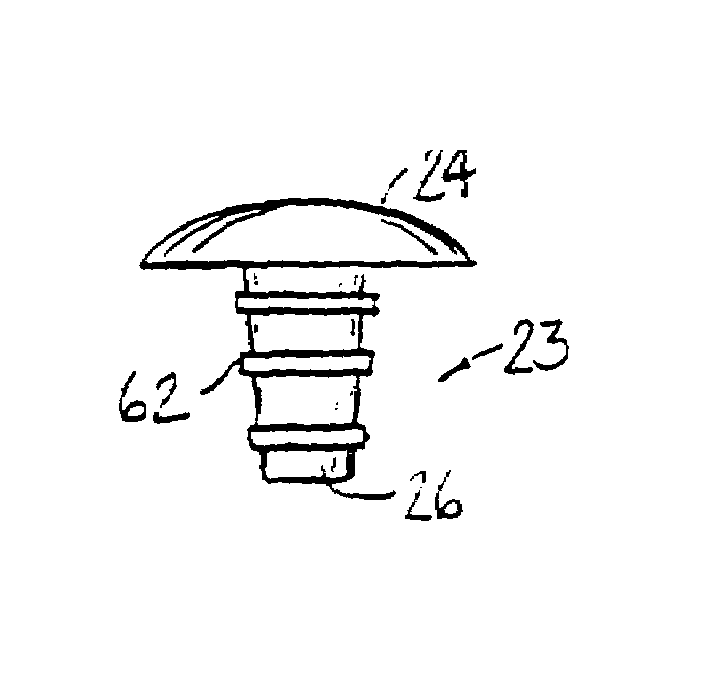
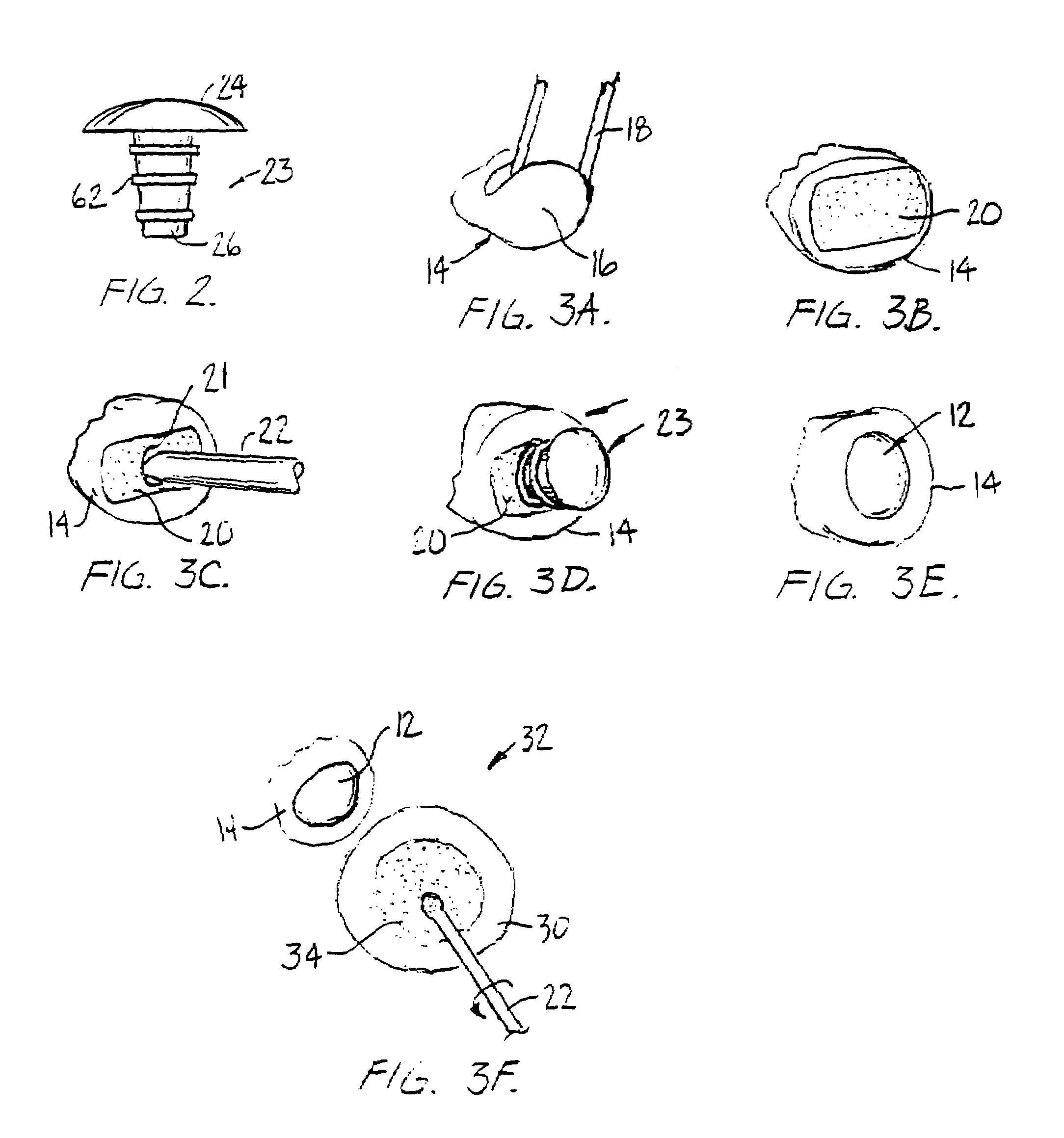
InactiveUS7128763B1Stimulate and promote formationKeep movingJoint implantsArticular surfacesFibrocartilage
A method for treating a non-weight bearing arthritic joint involves resecting at least one of the opposed joint surfaces to expose a cancellous bone surface. A bioresorbable implant is mounted to one of the joint surfaces so that the resected joint surface rubs against the surface of the implant. This causes the fibroblast to change into fibrocartilage at the resected bone surface as the implant is resorbed thereby effectively replacing the implant with fibrocartilage during such resorption.
Owner:BLATT GERALD
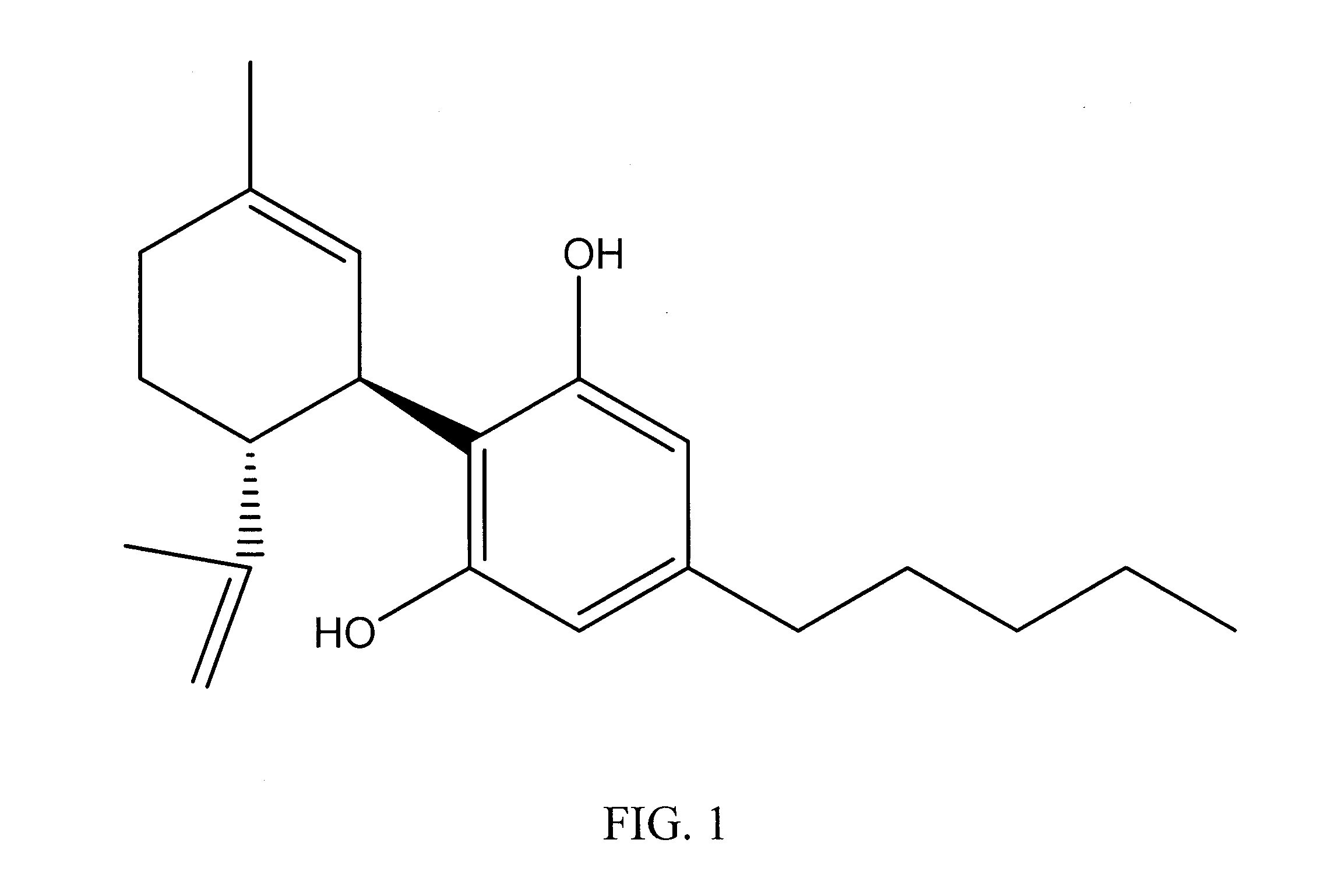
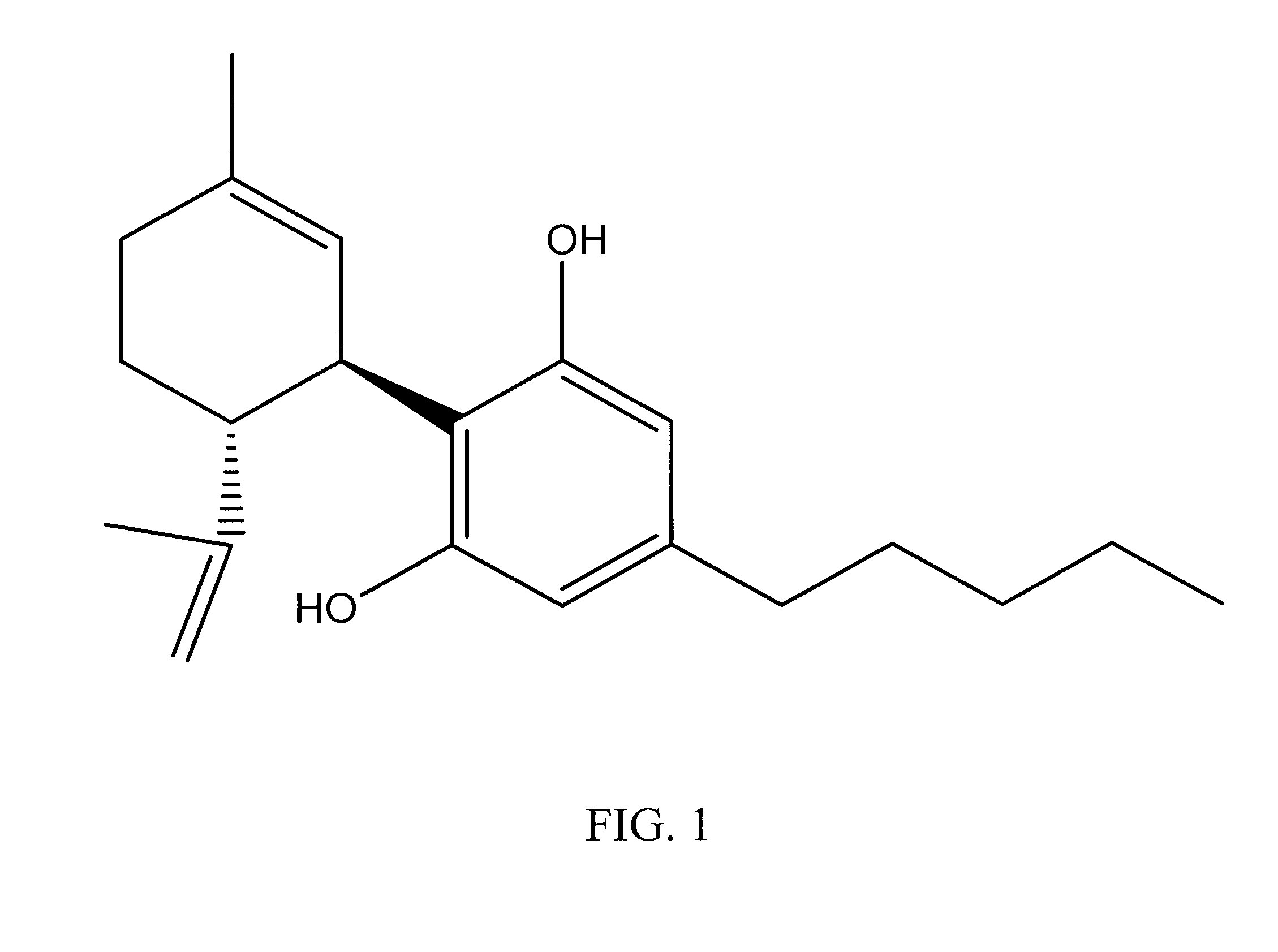
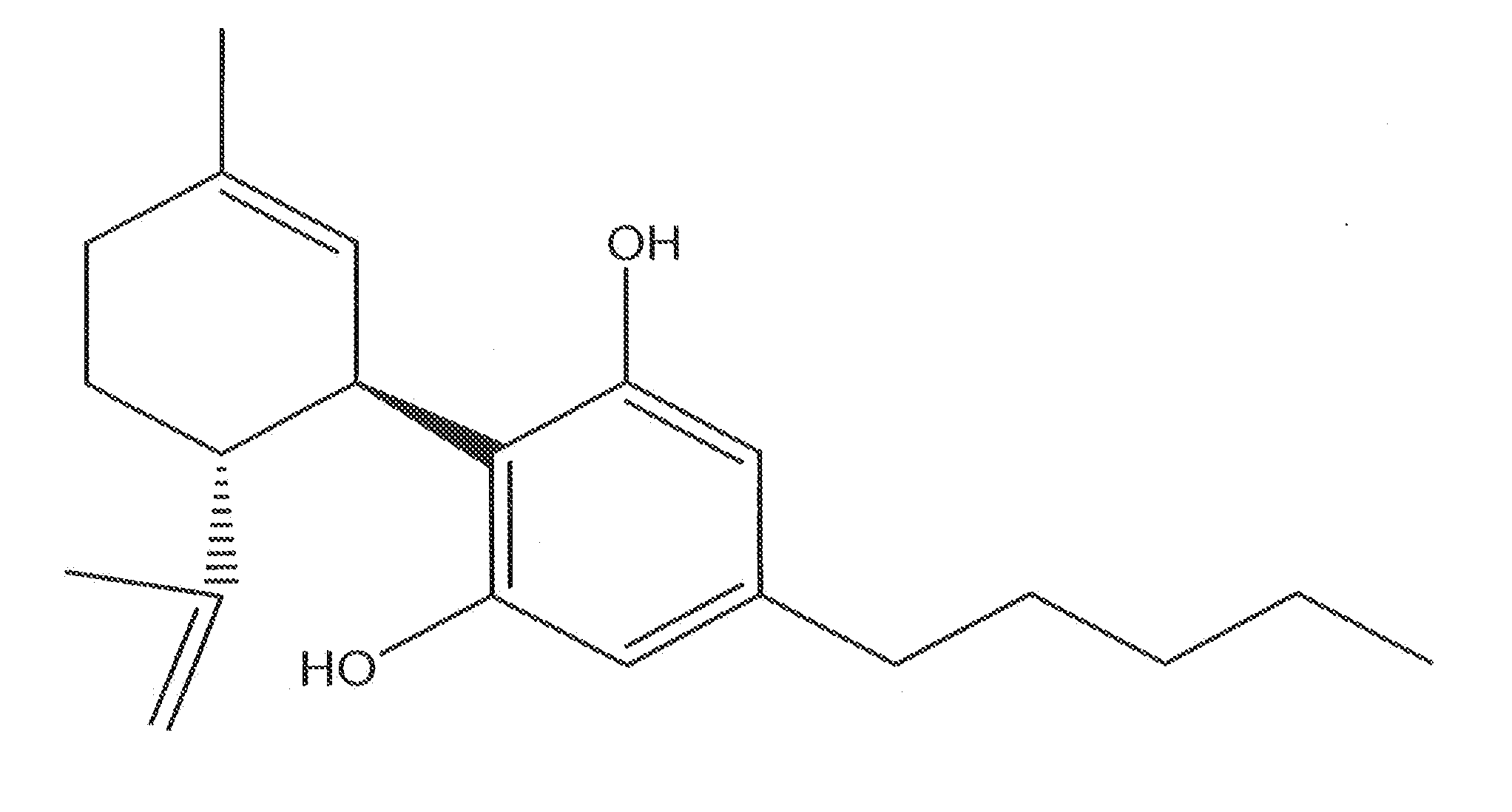
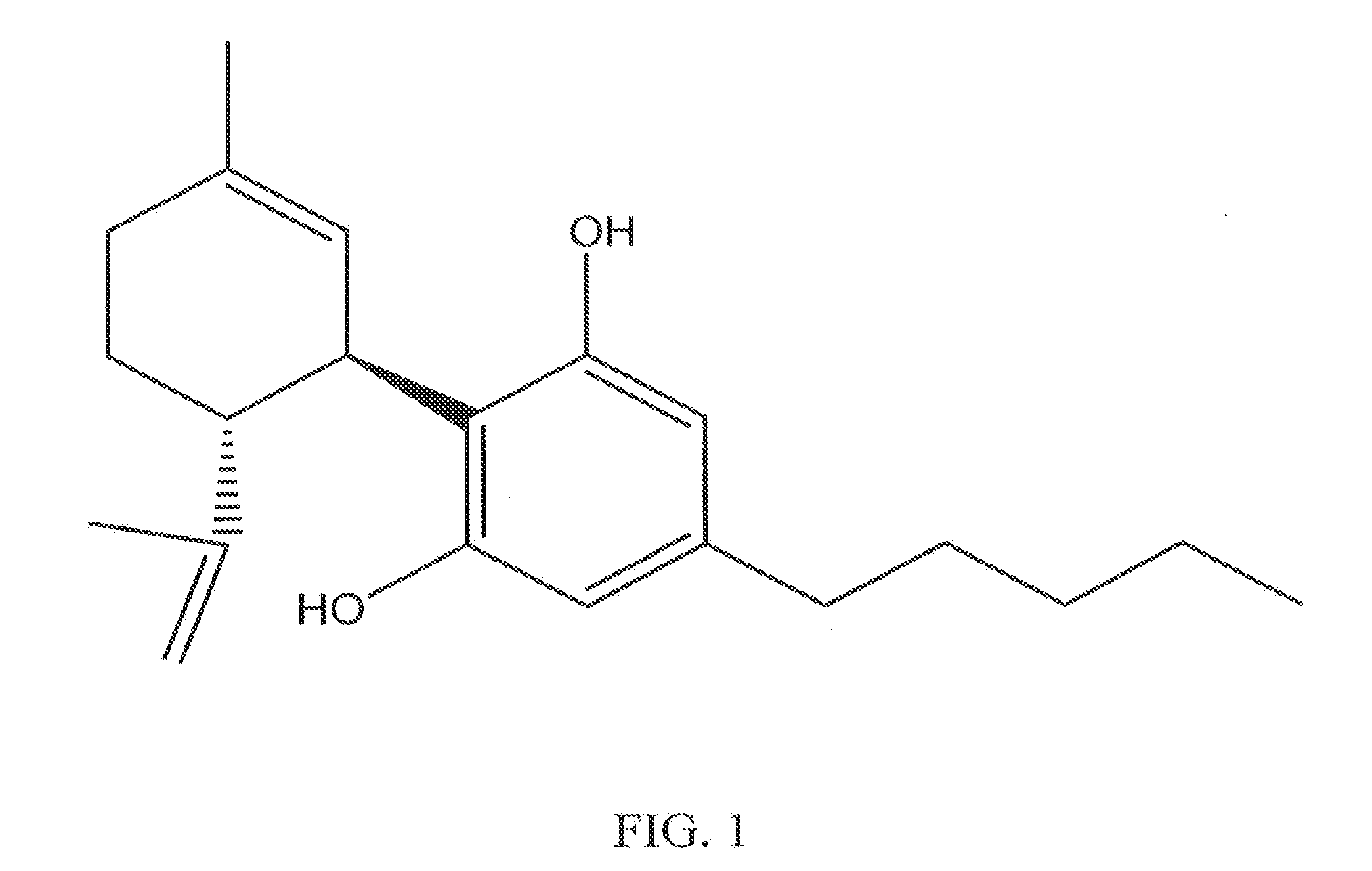
Cannabinoid-Containing Compositions and Methods for Their Use
InactiveUS20120202891A1Improve clinical efficacyReduce inflammationBiocideHydroxy compound active ingredientsInjury causeCannabis
This invention relates to cannabinoid-containing compositions, particularly cannabinoid-containing gel formulations and methods for the treatment of traumatic injury, e.g., strains, sprains and contusions, and disease conditions, e.g., arthritis, particularly osteoarthritis. The methods involve topically applying a cannabinoid or a cannabinoid-containing composition to a subject's skin near, or distant from, the area of the injury or the area affected by the disease condition, e.g., an arthritic joint. The cannabinoid-containing composition is preferably a pharmaceutically acceptable gel containing a therapeutically effective amount of a cannabinoid sufficient to alleviate the symptoms associate with the injury or disease condition.
Owner:KENTUCKY ECONOMIC DEV FINANCE AUTHORITY +1
Liposomal encapsulation of glycosaminoglycans for the treatment of arthritic joints
Owner:THOMPSON JONATHAN +1
Bracing and electrostimulation for arthritis
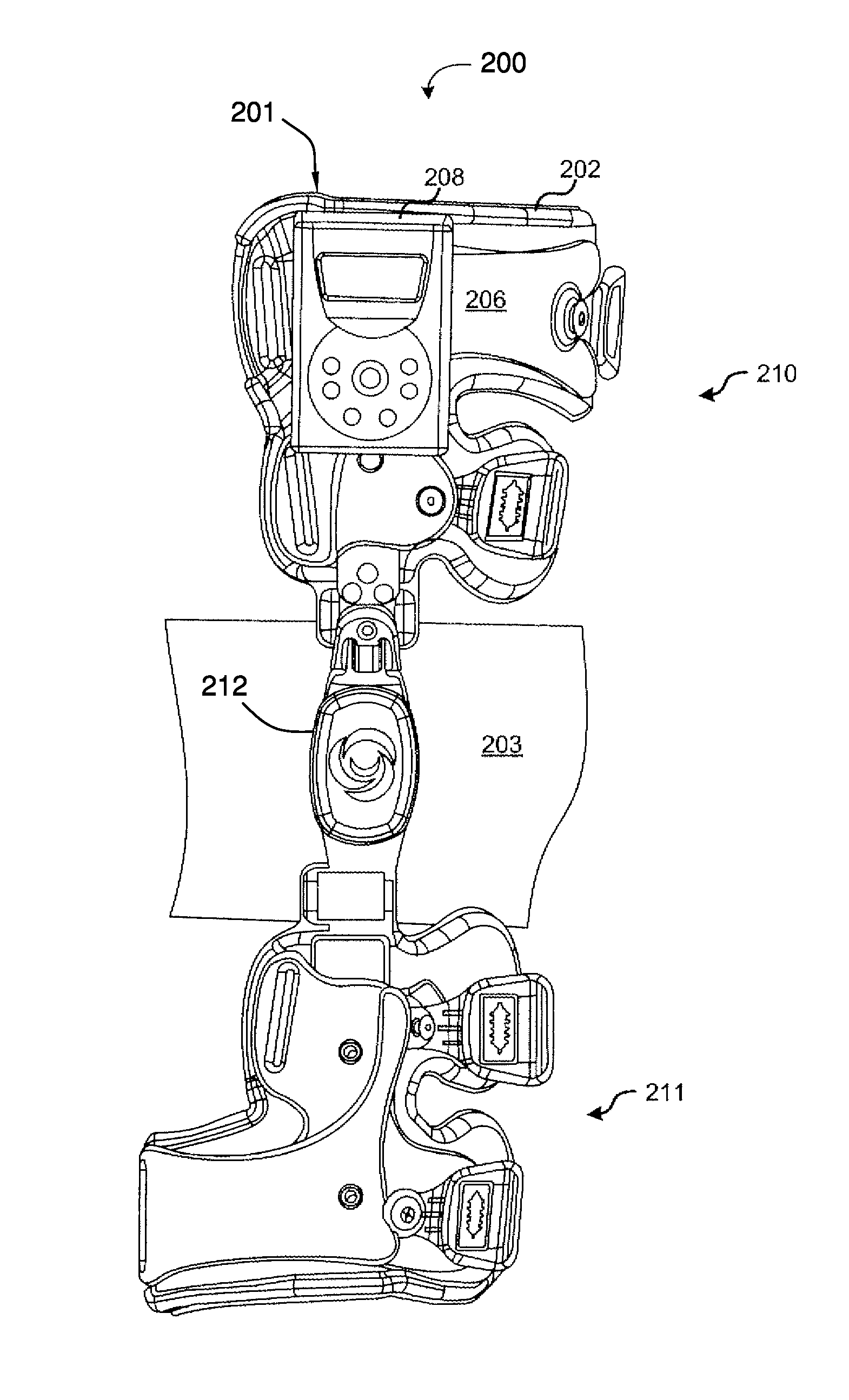
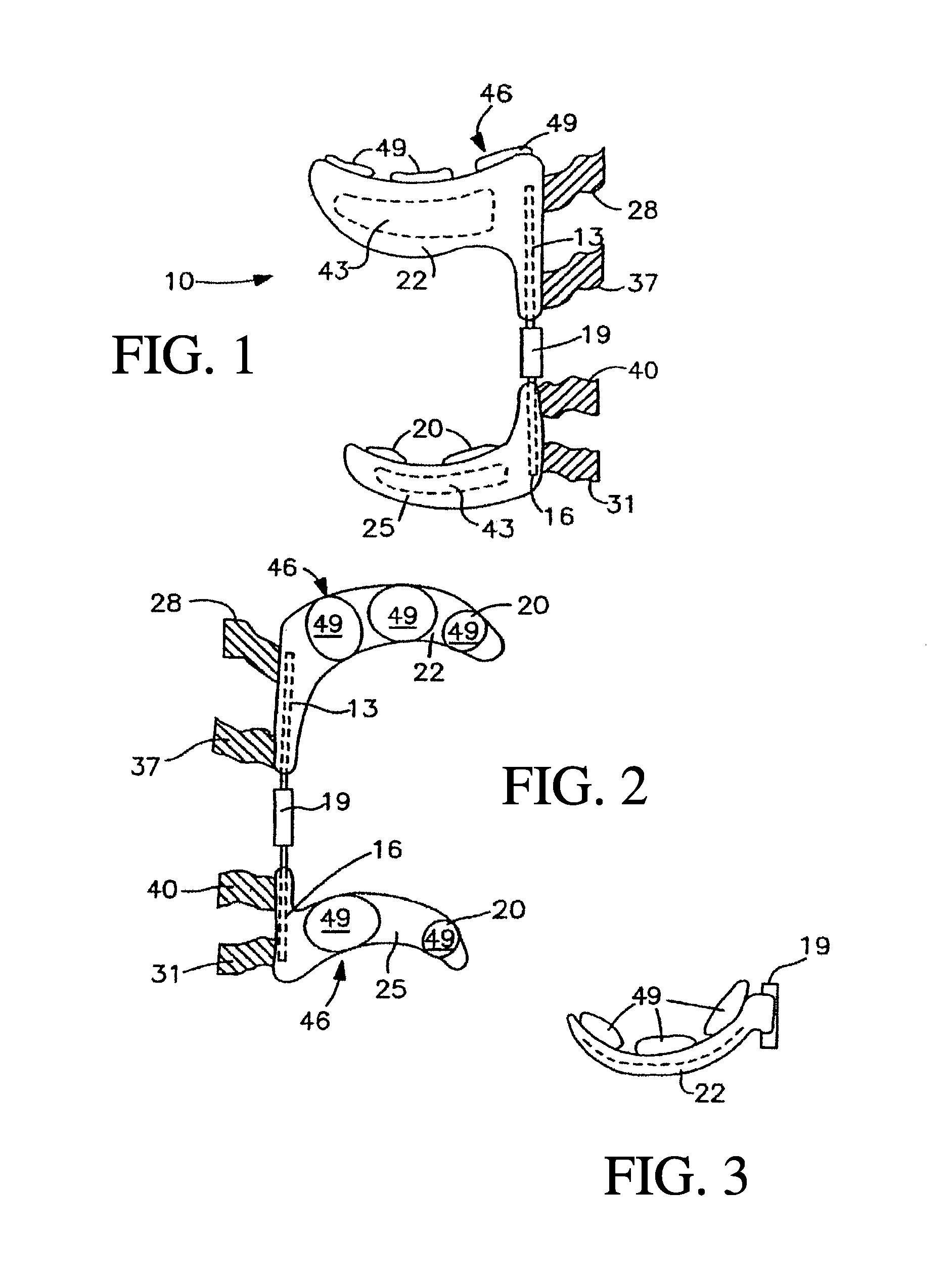
ActiveUS20100262052A1Function increaseOptimized joint treatmentElectrotherapyNon-surgical orthopedic devicesArthritic jointEngineering
A system for treating arthritis includes at least one joint stabilizing assembly for providing relief from arthritis. At least one signal transmission element engagement member is operatively connected to the joint stabilizing assembly for connecting a signal transmission element. At least one signal transmission element is supported by the at least one signal transmission element engagement member. An electrostimulation unit, electrically connected to the signal transmission element produces a signal for improving the overall function of an arthritic joint. The electrostimulation unit used in conjunction with the joint stabilizing assembly provides a synergistic effect, which results in optimized joint treatment versus using either the joint stabilizing assembly or electrostimulation unit alone.
Owner:VISION QUEST IND VQ ORTHOCARE
Bracing and electrostimulation for arthritis
InactiveUS20110288611A1Function increaseElectrotherapyNon-surgical orthopedic devicesElectricityArthritic joint
A system for treating arthritis includes at least one joint stabilizing assembly for providing relief from arthritis. At least one signal transmission element engagement member is operatively connected to the joint stabilizing assembly for connecting a signal transmission element. At least one signal transmission element is supported by the at least one signal transmission element engagement member. An electrostimulation unit, electrically connected to the signal transmission element produces a signal for improving the overall function of an arthritic joint. The electrostimulation unit used in conjunction with the joint stabilizing assembly for greater than 150 hours provides a synergistic effect, which results in optimized joint treatment versus using either the joint stabilizing assembly or electrostimulation unit alone.
Owner:VISION QUEST IND VQ ORTHOCARE
Cannabinoid-Containing Compositions and Methods for Their Use
InactiveUS20090247619A1Improve clinical efficacyReduce inflammationBiocideHydroxy compound active ingredientsArthritic jointTraumatic injury
This invention relates to cannabinoid-containing compositions, particularly cannabinoid-containing gel formulations and methods for the treatment of traumatic injury, e.g., strains, sprains and contusions, and disease conditions, e.g., arthritis, particularly osteoarthritis. The methods involve topically applying a cannabinoid or a cannabinoid-containing composition to a subject's skin near, or distant from, the area of the injury or the area affected by the disease condition, e.g., an arthritic joint. The cannabinoid-containing composition is preferably a pharmaceutically acceptable gel containing a therapeutically effective amount of a cannabinoid sufficient to alleviate the symptoms associate with the injury or disease condition.
Owner:KENTUCKY ECONOMIC DEV FINANCE AUTHORITY
Treatment of amelioration of arthritic joint pain
InactiveUS20100113862A1Easy to useMore comfortableElectrotherapyMagnetotherapy using coils/electromagnetsPower flowArthritic pains
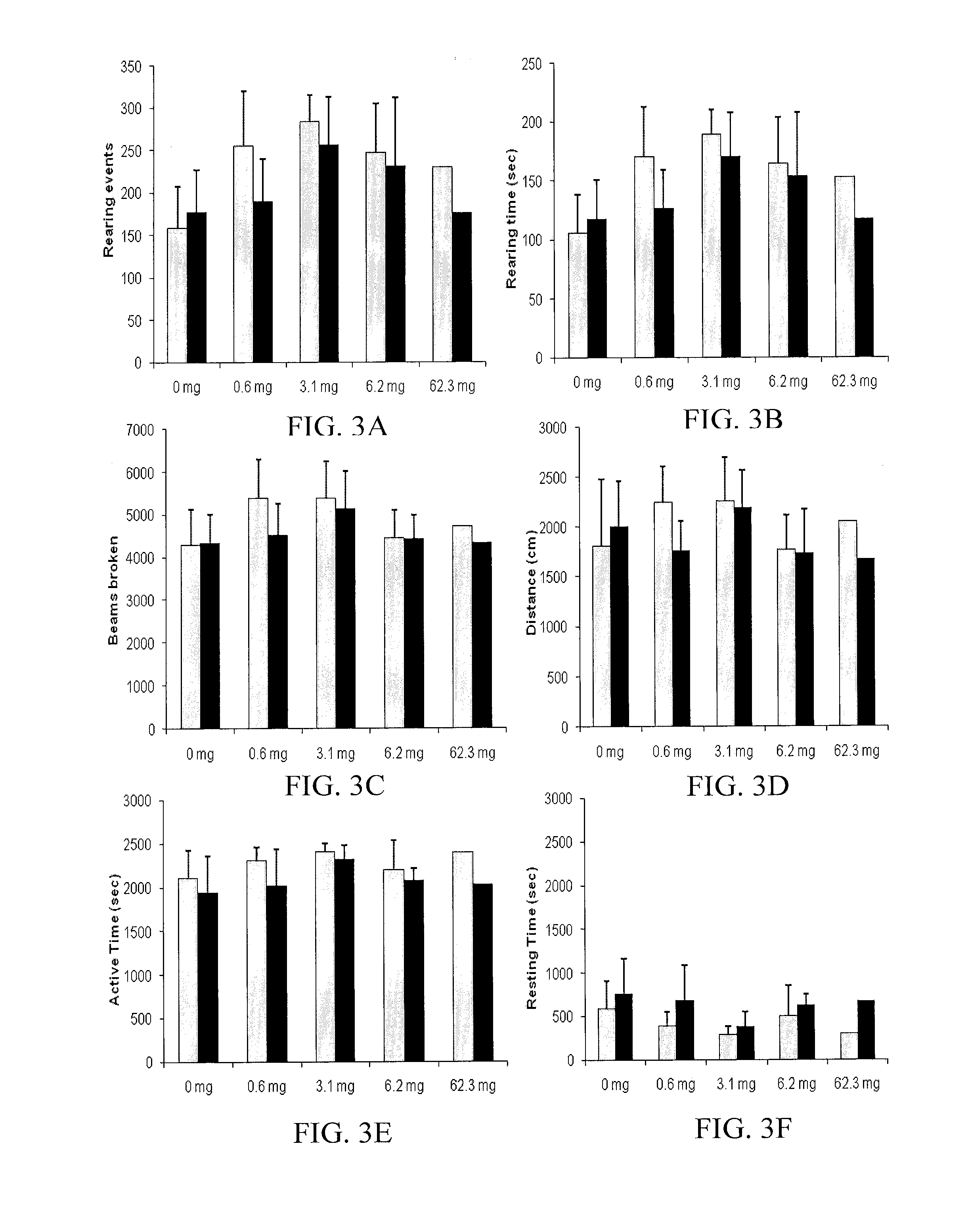
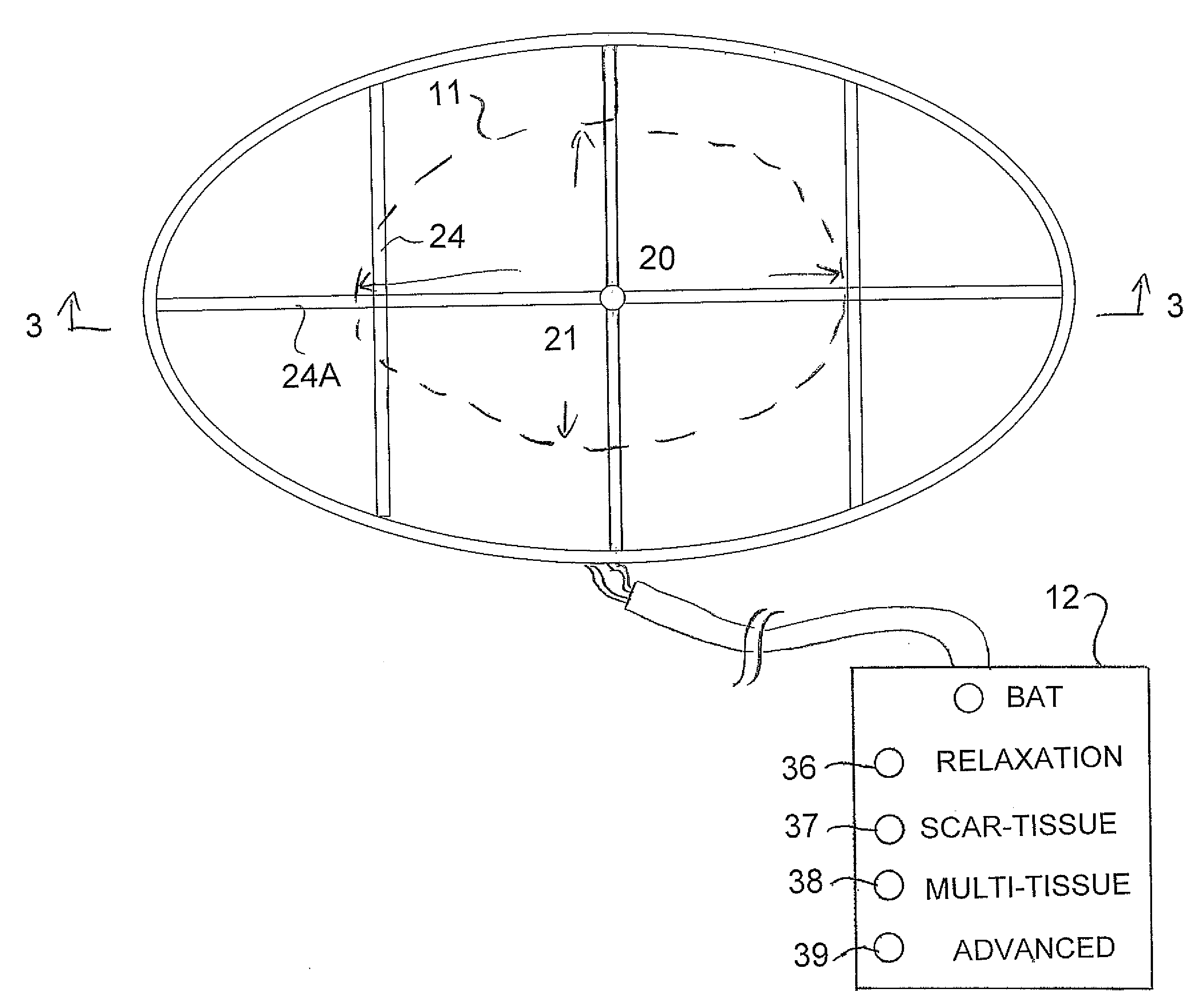
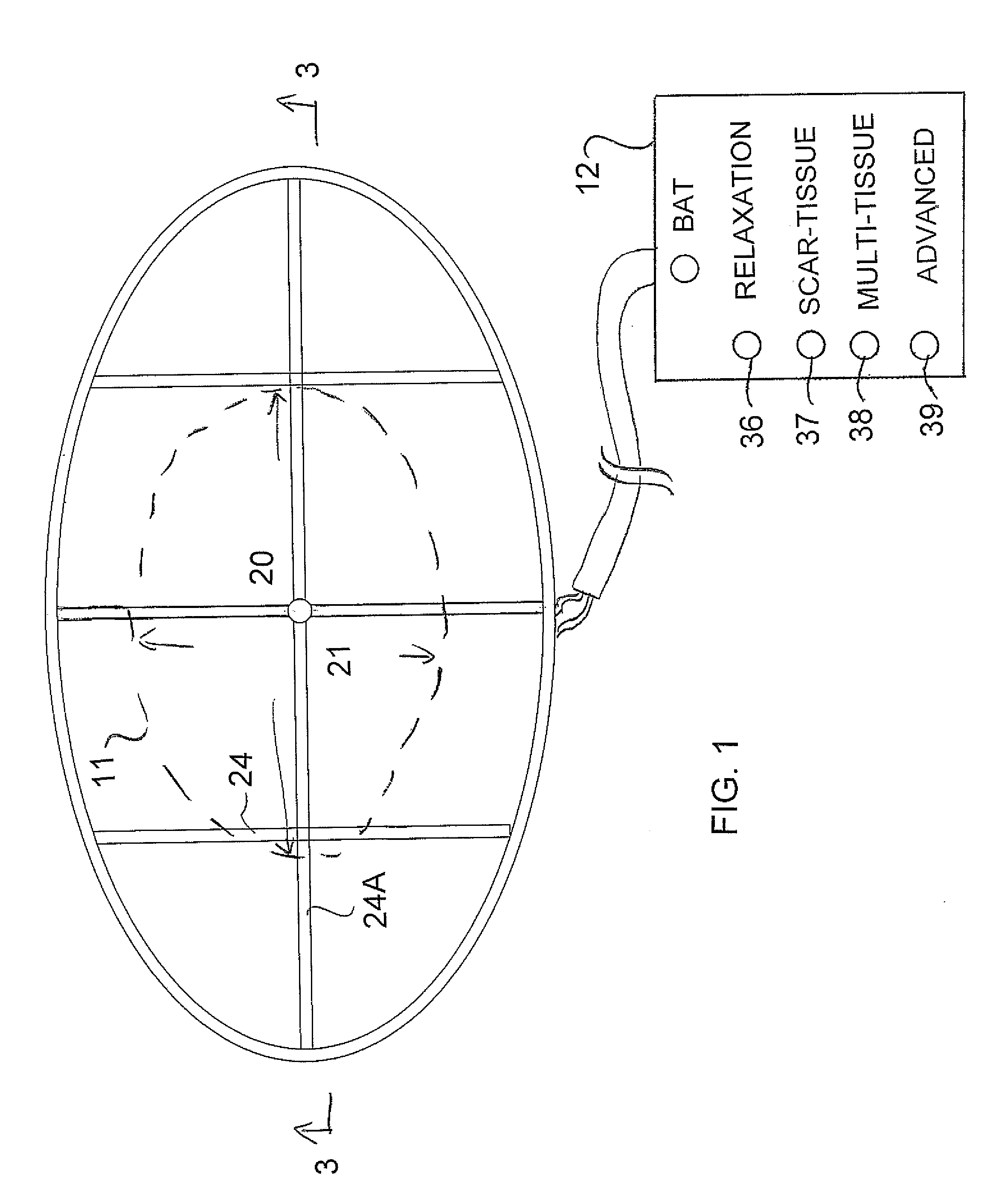
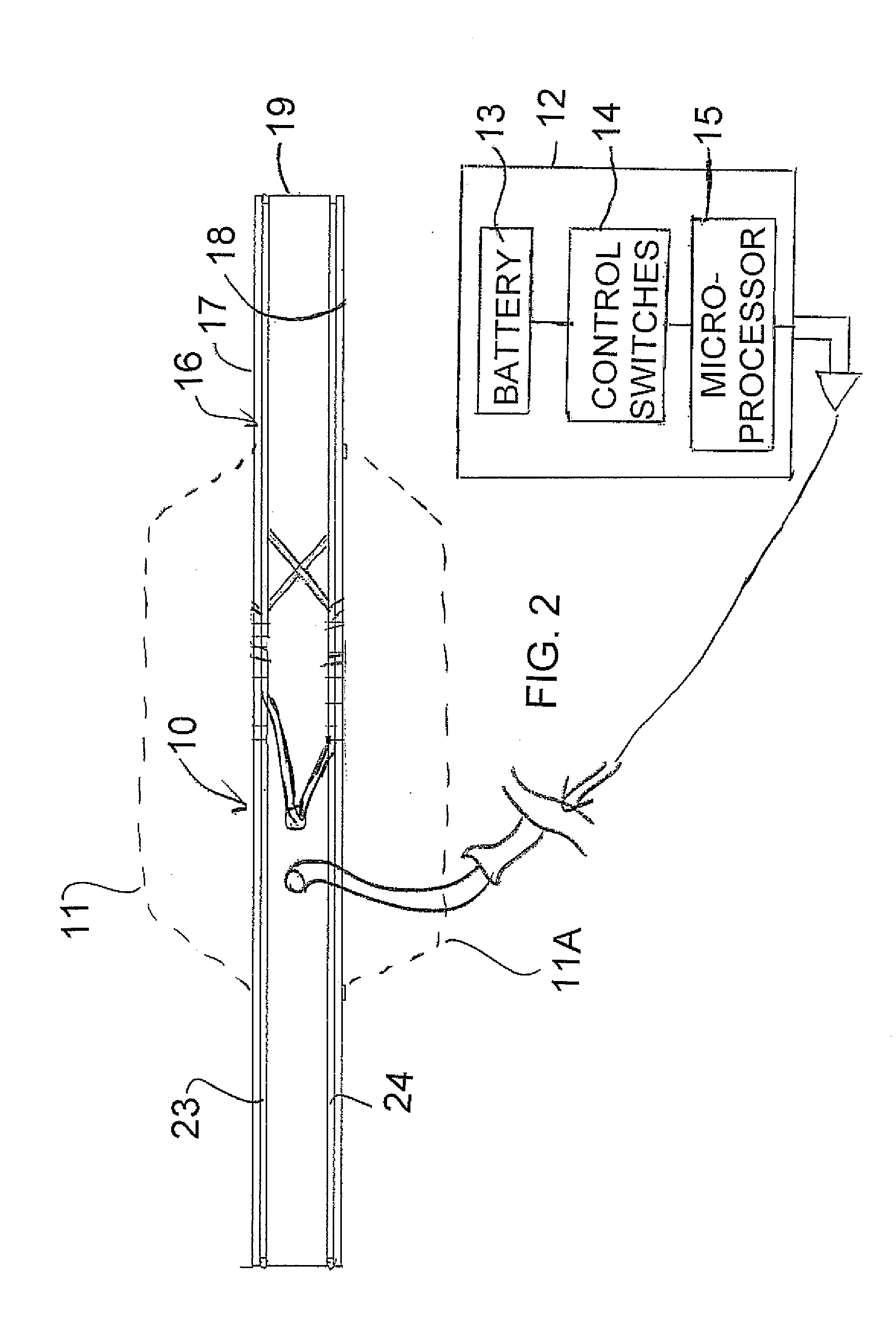
An apparatus for treatment of pain includes a coil formed by a generally flat elliptical base member wrapped by wire in two separated grooves spaced axially along the outer surface and a portable battery powered generator unit for generating a current such that the electromagnetic field has a required magnitude of magnetic flux density in the range 1×104−6 gauss to 1×10−12 gauss at a frequency of between 0.01 and 100 Hz. The base member of the coil is covered by a flexible fabric covering material with a padding layer on each side. The coil is elliptical in plan to generate a field of the required magnitude emitted outwardly to one side in an area of the order of 10 inches by 5 inches at a depth of 2 inches. The generator is controlled wholly by simple switches which only select from a plurality of treatment options.
Owner:KOTOWICH ALAN W J +1
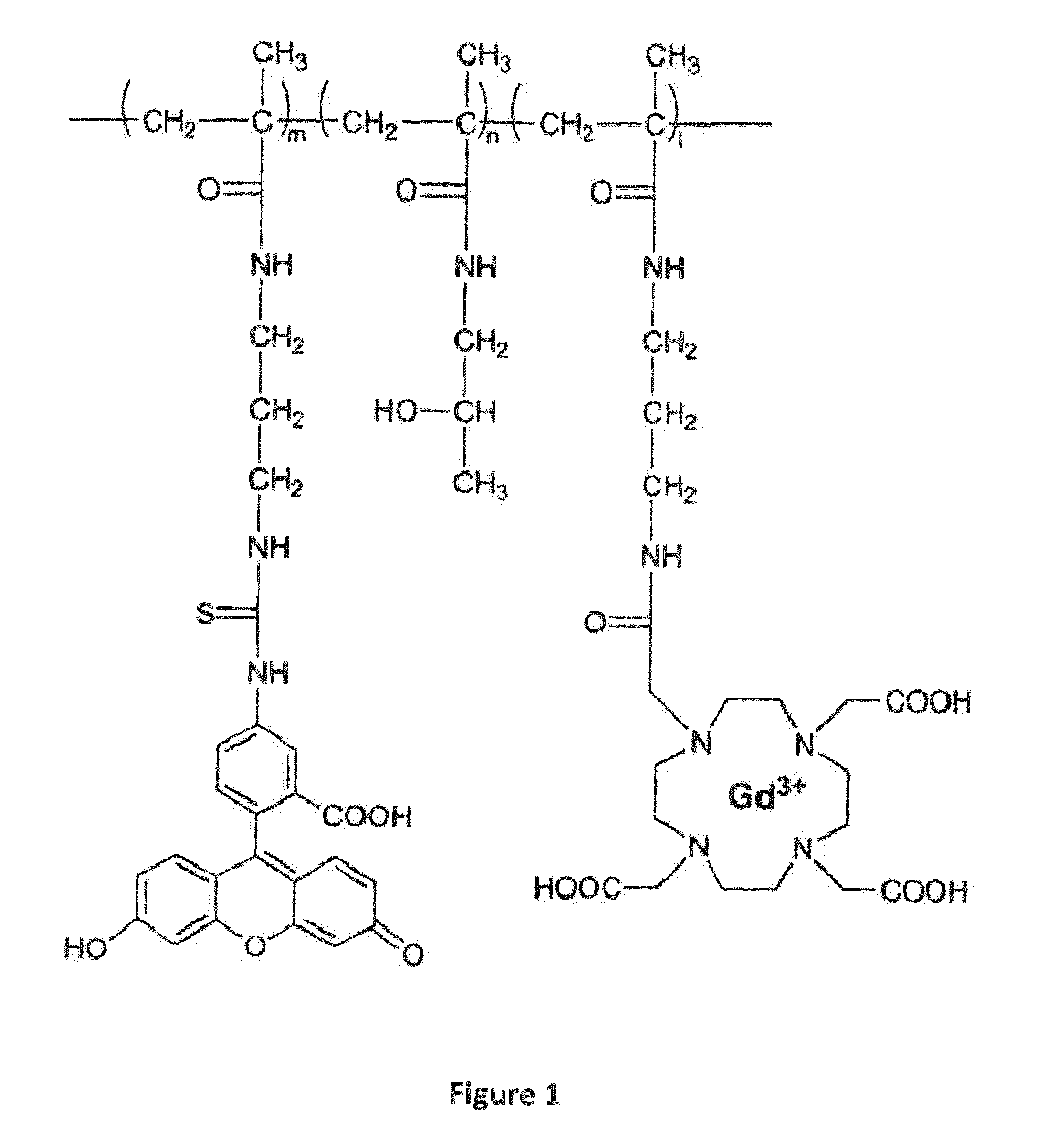
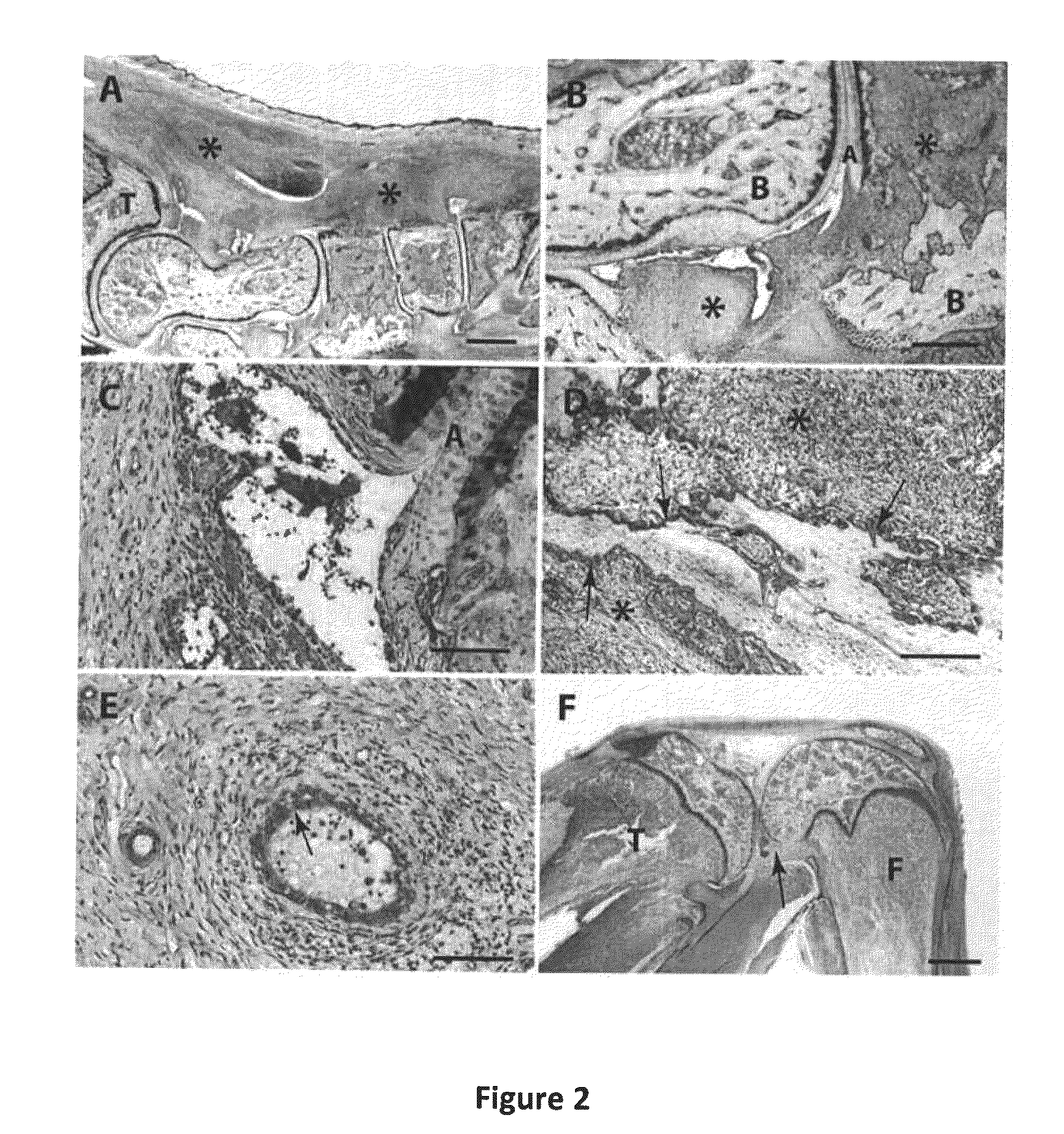
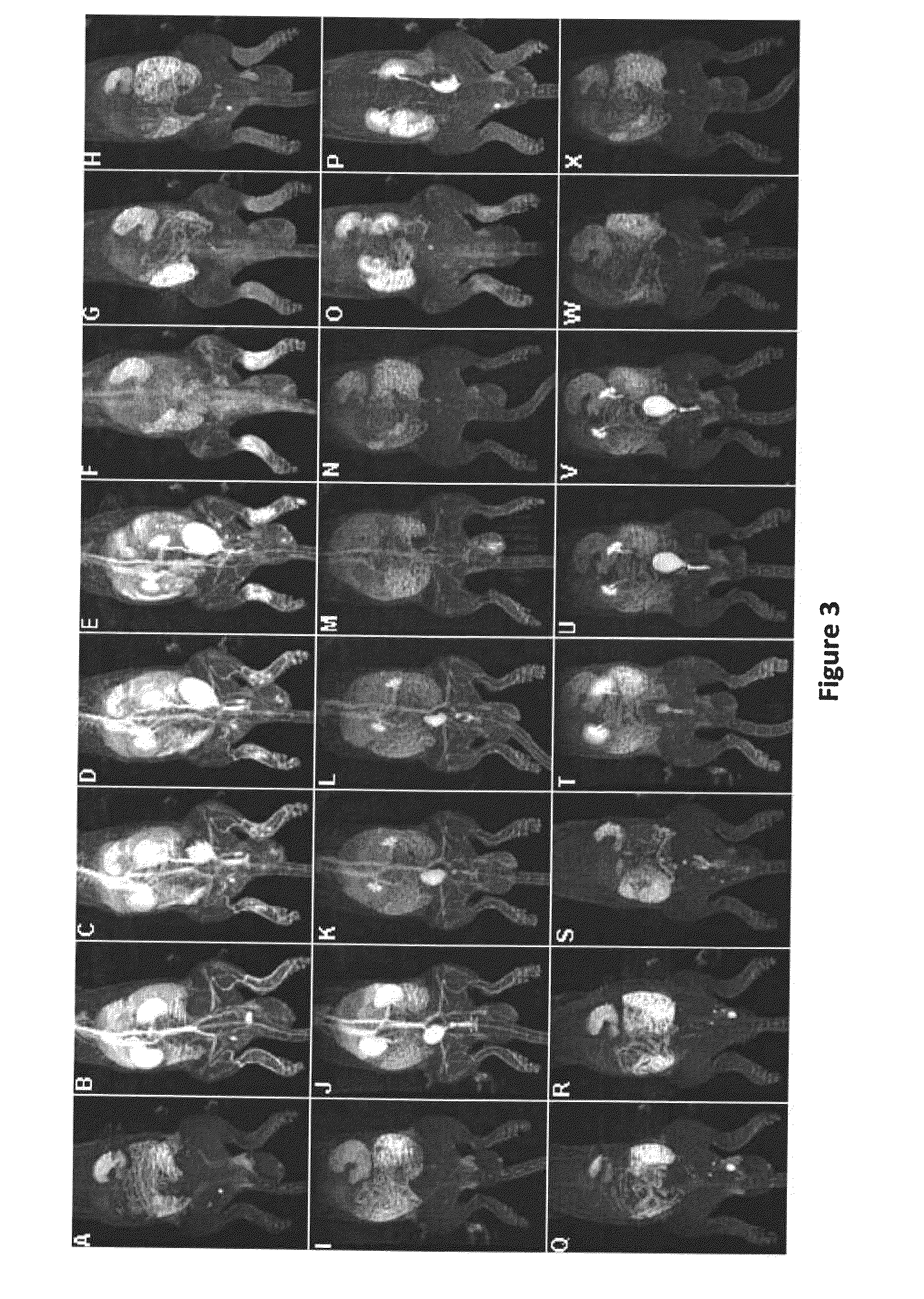
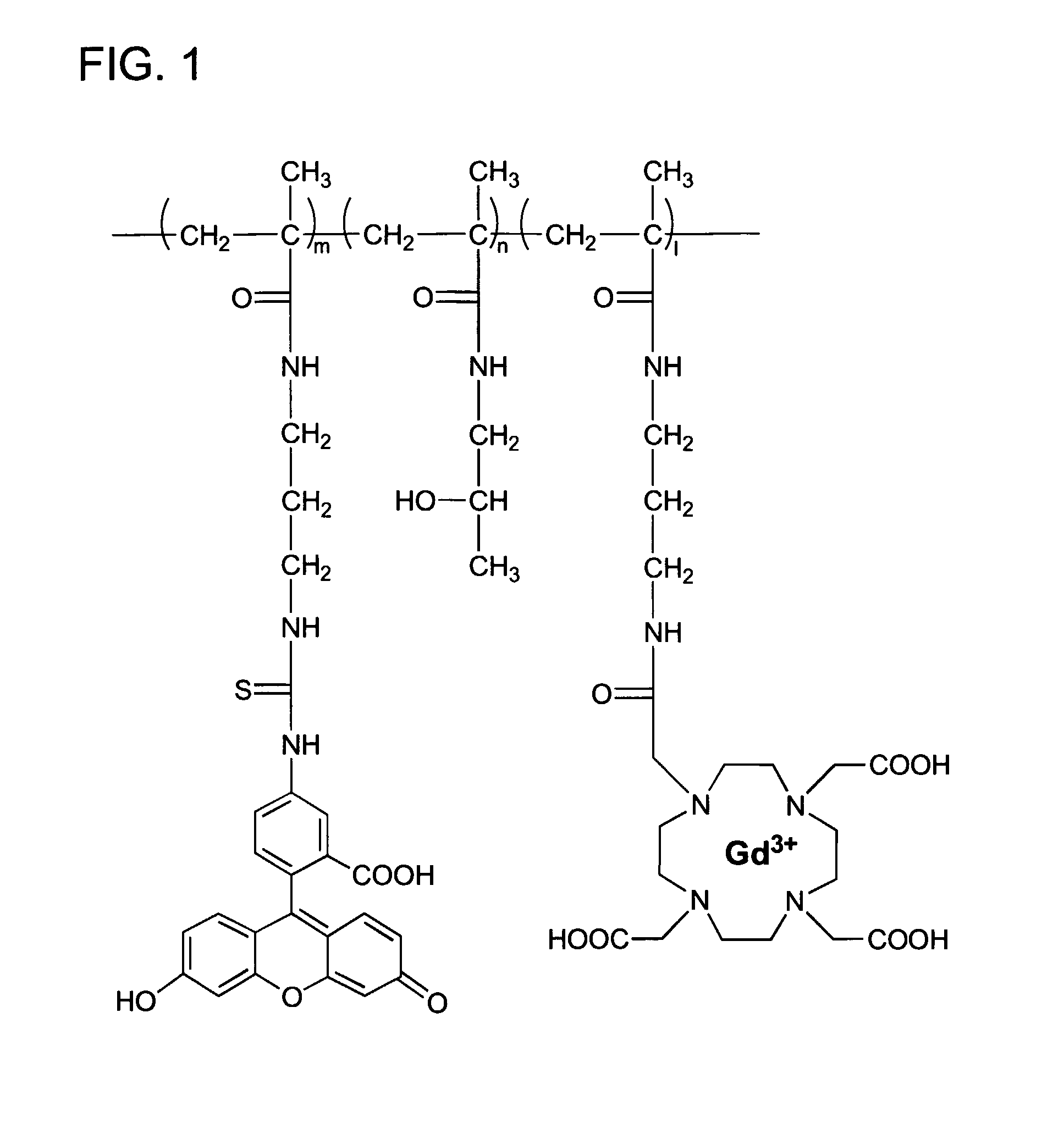
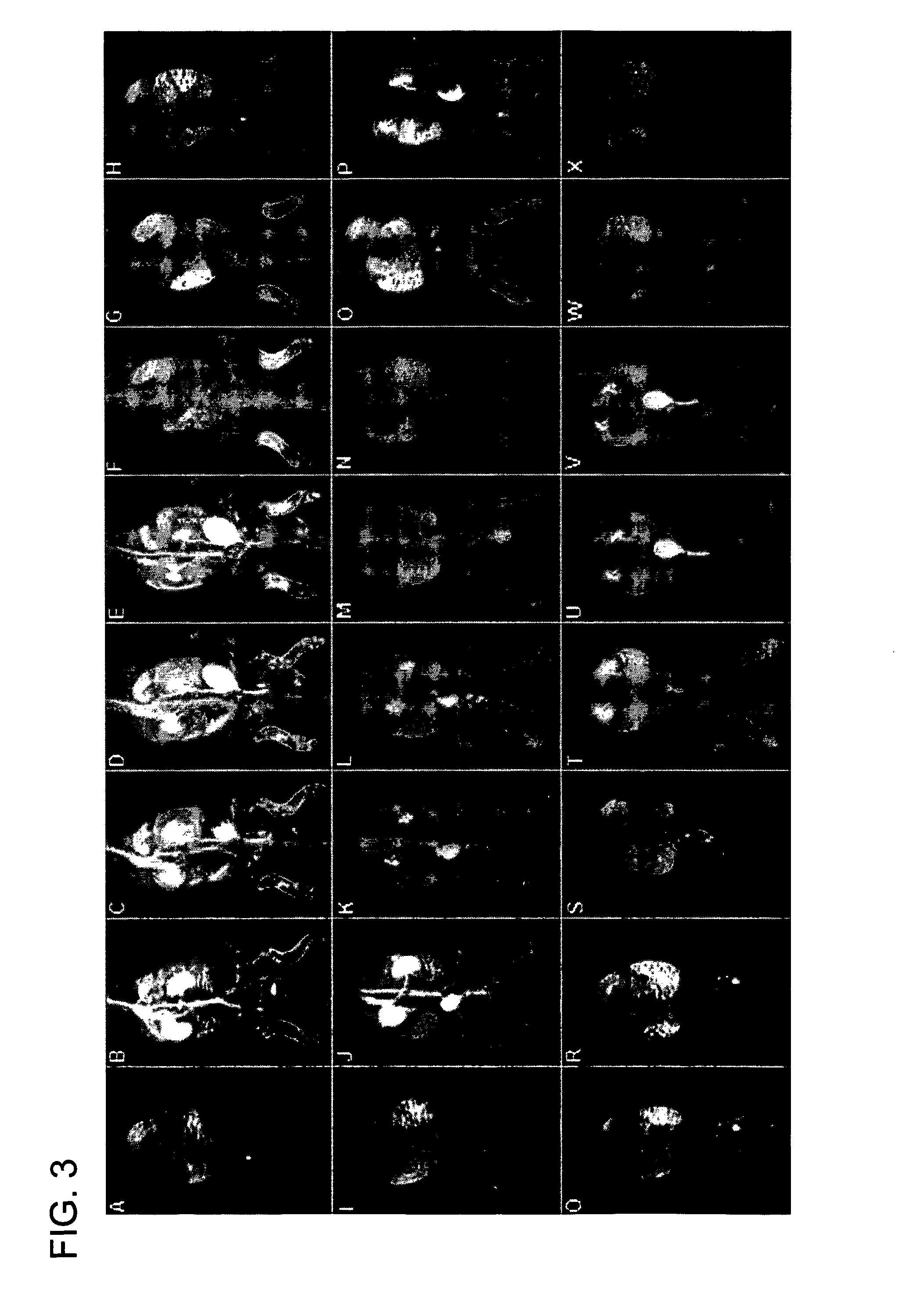
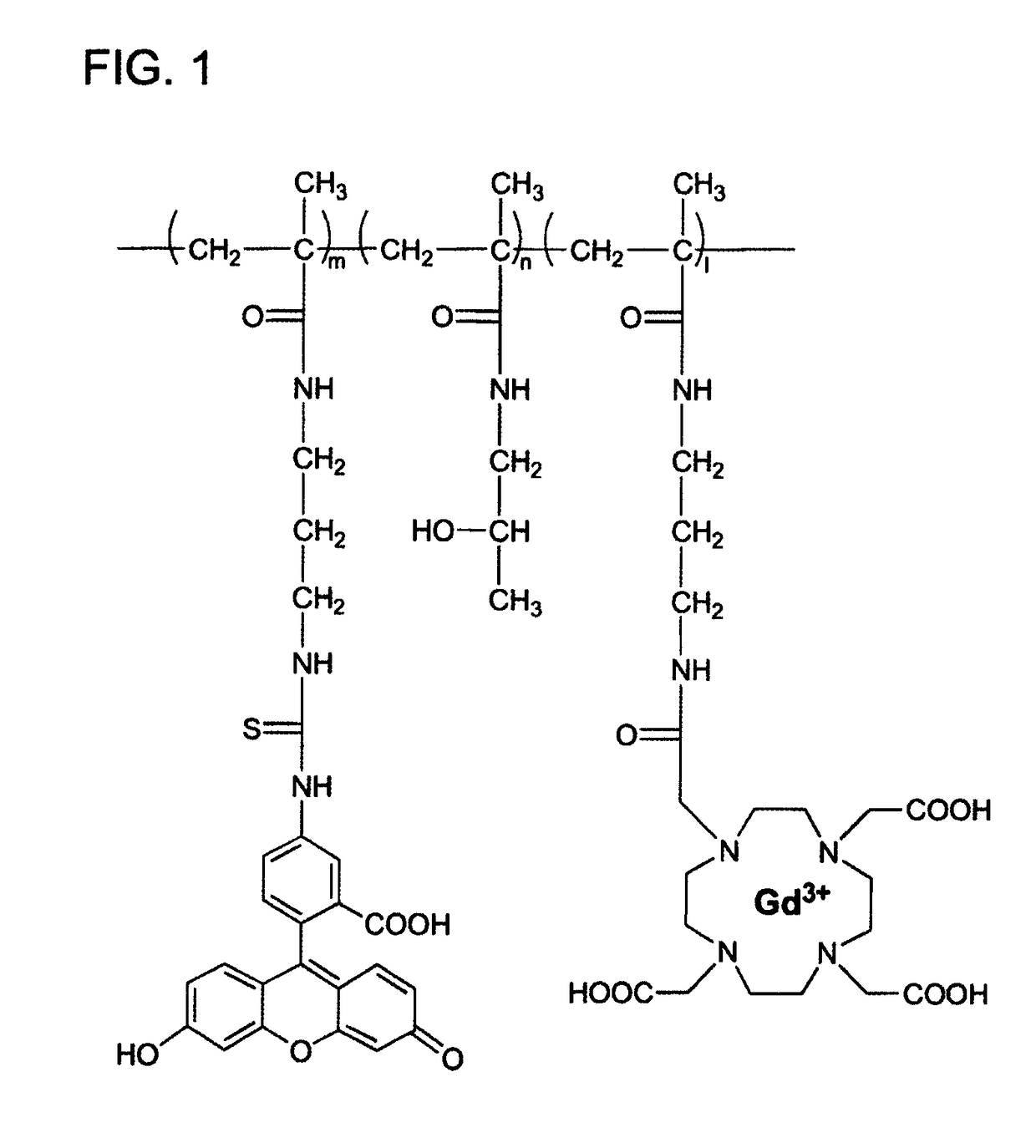
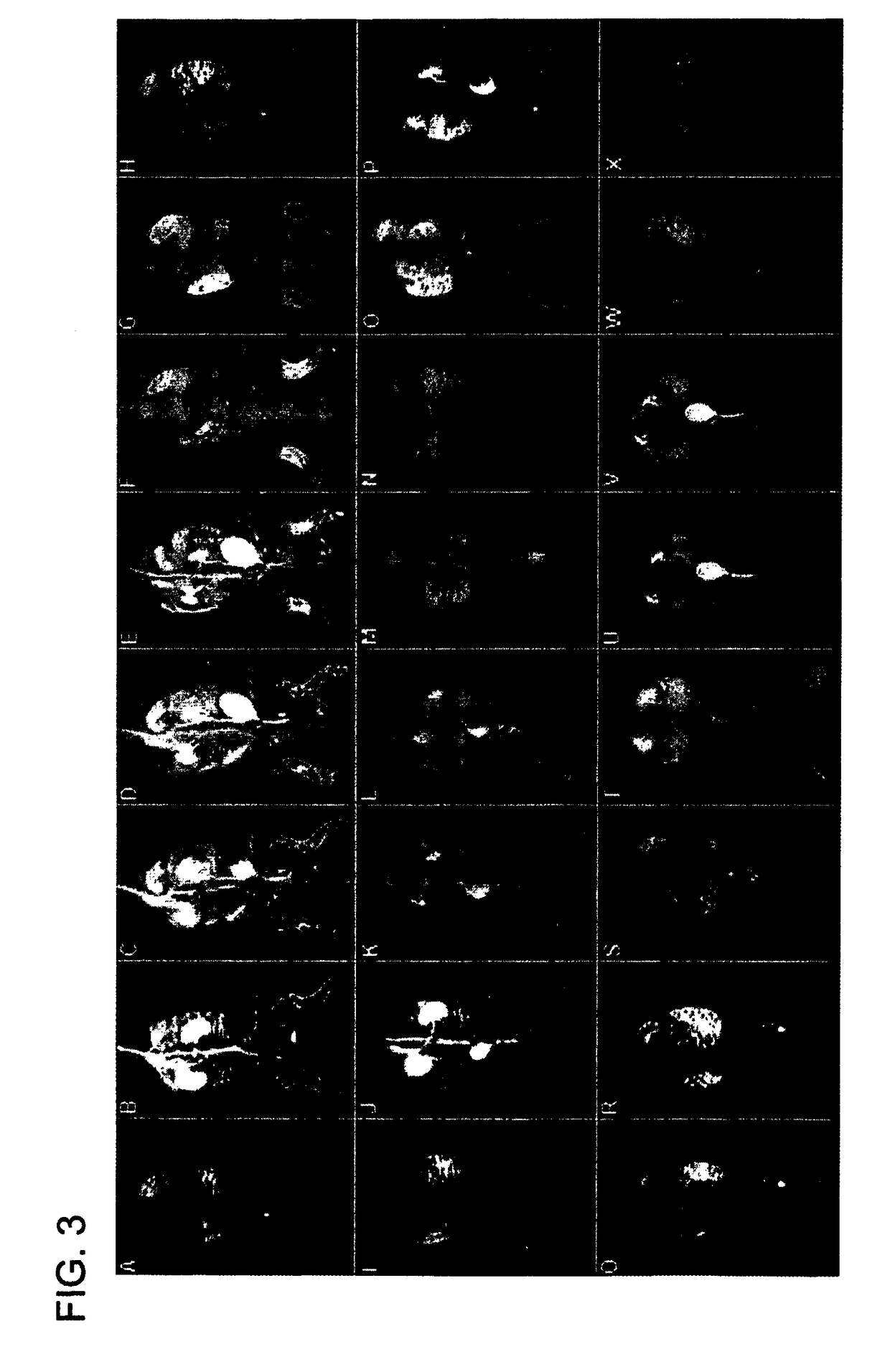
Macromolecular Delivery Systems for Non-Invasive Imaging, Evaluation and Treatment of Arthritis and Other Inflammatory Diseases
InactiveUS20090311182A1High retention rateGood treatment effectOrganic active ingredientsPeptide/protein ingredientsDiseaseNon invasive
This invention relates to biotechnology, more particularly, to water-soluble polymeric delivery systems for the imaging, evaluation and / or treatment of rheumatoid arthritis and other inflammatory diseases. Using modern MR imaging techniques, the specific accumulation of macromolecules in arthritic joints in adjuvant-induced arthritis in rats is demonstrated. The strong correlation between the uptake and retention of the MR contrast agent labeled polymer with histopathological features of inflammation and local tissue damage demonstrates the practical applications of the macromolecular delivery system of the invention.
Owner:BOARD OF RGT UNIV OF NEBRASKA
Cannabinoid-Containing Compositions and Methods for Their Use
InactiveUS20120202892A1Relief of inflammatory painControl rateBiocideHydroxy compound active ingredientsInjury causeCannabis
This invention relates to cannabinoid-containing compositions, particularly cannabinoid-containing gel formulations and methods for the treatment of traumatic injury, e.g., strains, sprains and contusions, and disease conditions, e.g., arthritis, particularly osteoarthritis. The methods involve topically applying a cannabinoid or a cannabinoid-containing composition to a subject's skin near, or distant from, the area of the injury or the area affected by the disease condition, e.g., an arthritic joint. The cannabinoid-containing composition is preferably a pharmaceutically acceptable gel containing a therapeutically effective amount of a cannabinoid sufficient to alleviate the symptoms associate with the injury or disease condition.
Owner:KENTUCKY ECONOMIC DEV FINANCE AUTHORITY
Bracing and electrostimulation for arthritis
ActiveUS8936560B2Function increaseOptimized joint treatmentNon-surgical orthopedic devicesDiagnostic recording/measuringEnarthrosisElectricity
A system for treating arthritis includes at least one joint stabilizing assembly for providing relief from arthritis. At least one signal transmission element engagement member is operatively connected to the joint stabilizing assembly for connecting a signal transmission element. At least one signal transmission element is supported by the at least one signal transmission element engagement member. An electrostimulation unit, electrically connected to the signal transmission element produces a signal for improving the overall function of an arthritic joint. The electrostimulation unit used in conjunction with the joint stabilizing assembly provides a synergistic effect, which results in optimized joint treatment versus using either the joint stabilizing assembly or electrostimulation unit alone.
Owner:VISION QUEST IND VQ ORTHOCARE
Compositions and methods for the systemic treatment of arthritis
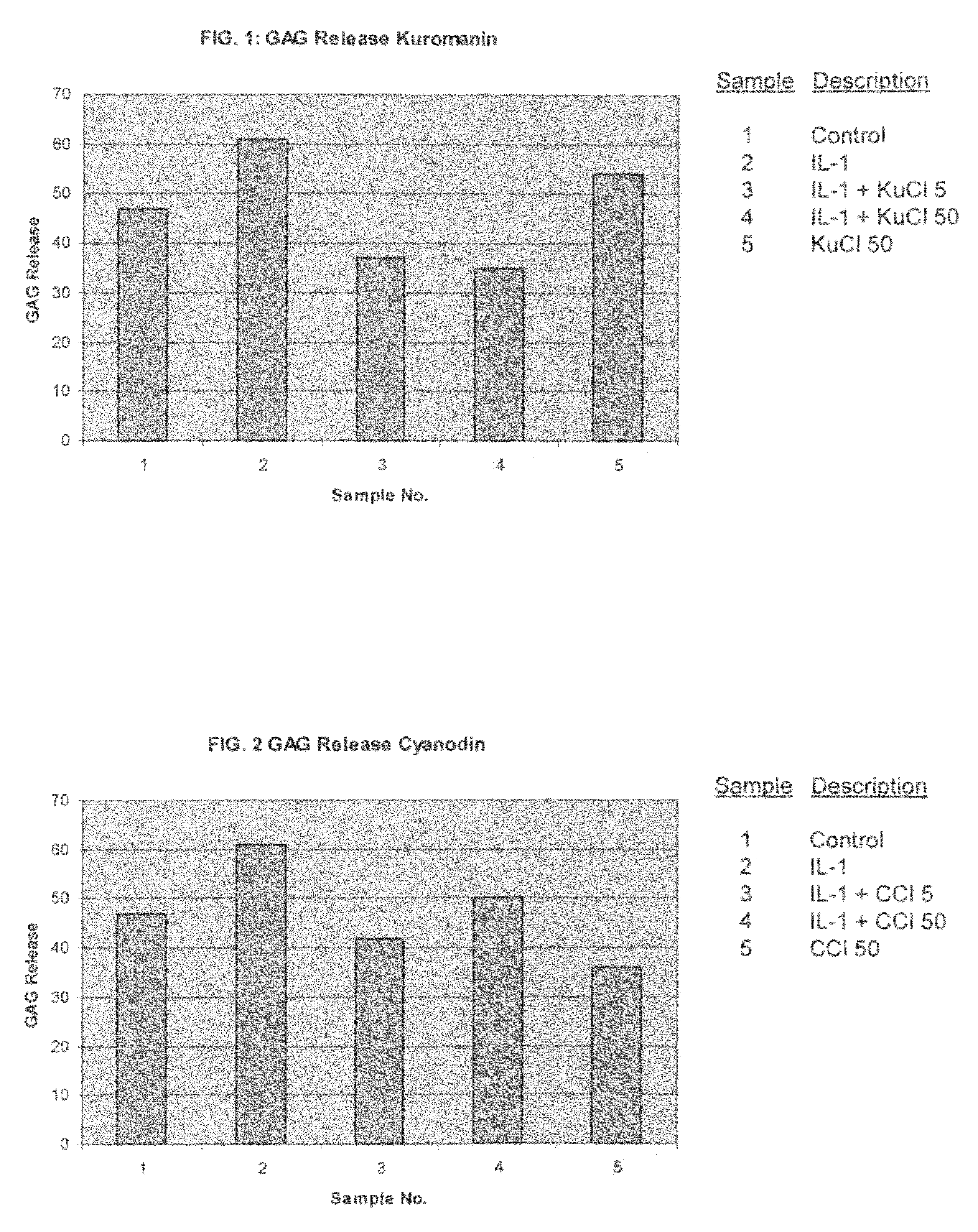
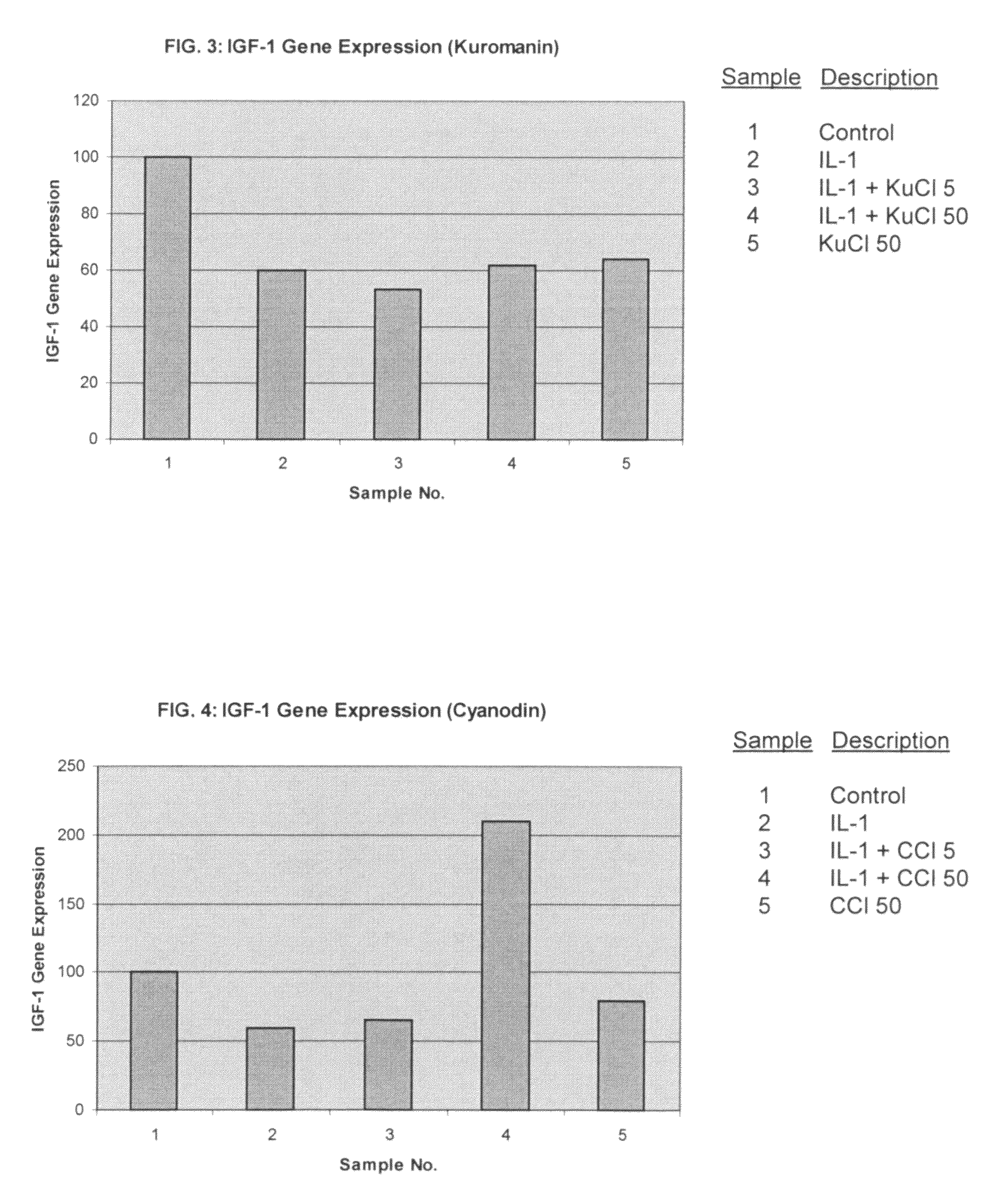
ActiveUS20050192241A1Reducing and blocking bioavailabilityEffective therapyPeptide/protein ingredientsAntipyreticArthritic jointBioavailability
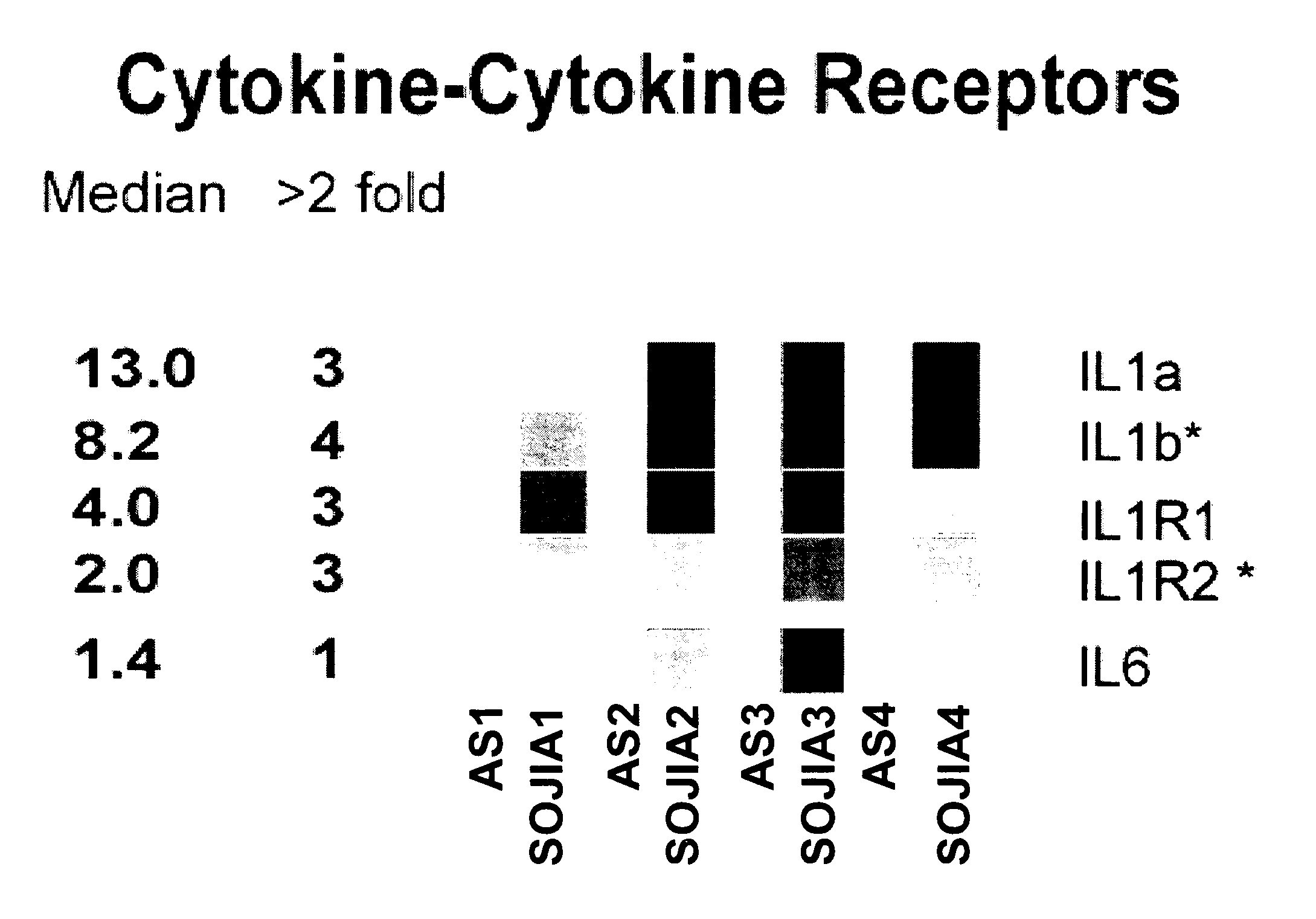
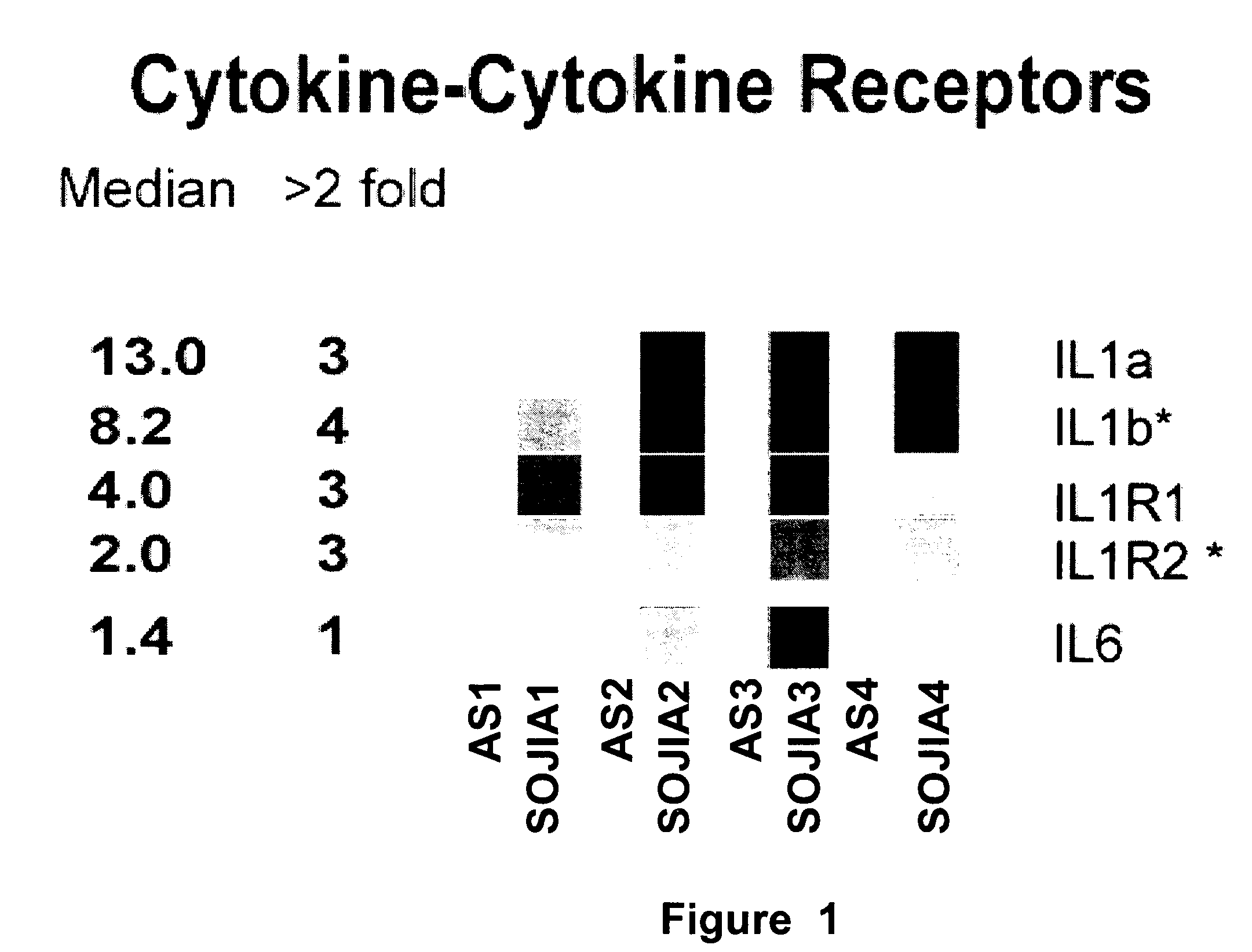
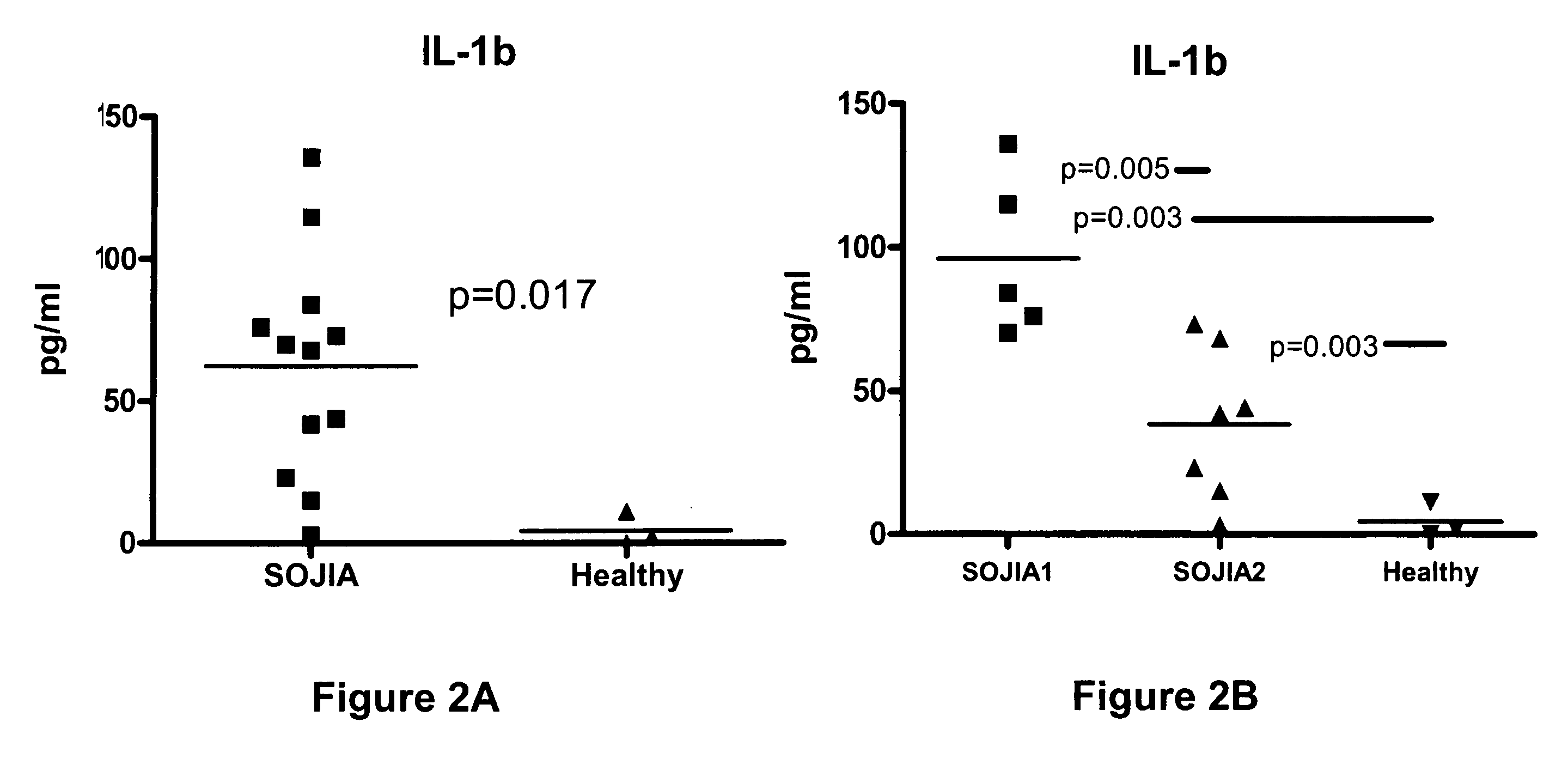
The present invention includes compositions and methods for treating arthritic joints found in patients with autoinflammation, e.g., systemic onset juvenile idiopathic arthritis, by administering at the site of inflammation a therapeutically effective amount of at least one agent that reduces or blocks the bioavailability of interleukin-1β.
Owner:BAYLOR RES INST
Macromolecular Delivery Systems for Non-Invasive Imaging, Evaluation and Treatment of Arthritis and Other Inflammatory Diseases
ActiveUS20080159959A1Powder deliveryHeavy metal active ingredientsMr contrast agentPathology diagnosis
This invention relates to biotechnology, more particularly, to water-soluble polymeric delivery systems for the imaging, evaluation and / or treatment of rheumatoid arthritis and other inflammatory diseases. Using modern MR imaging techniques, the specific accumulation of macromolecules in arthritic joints in adjuvant-induced arthritis in rats is demonstrated. The strong correlation between the uptake and retention of the MR contrast agent labeled polymer with histopathological features of inflammation and local tissue damage demonstrates the practical applications of the macromolecular delivery system of the invention.
Owner:UNIV OF UTAH RES FOUND
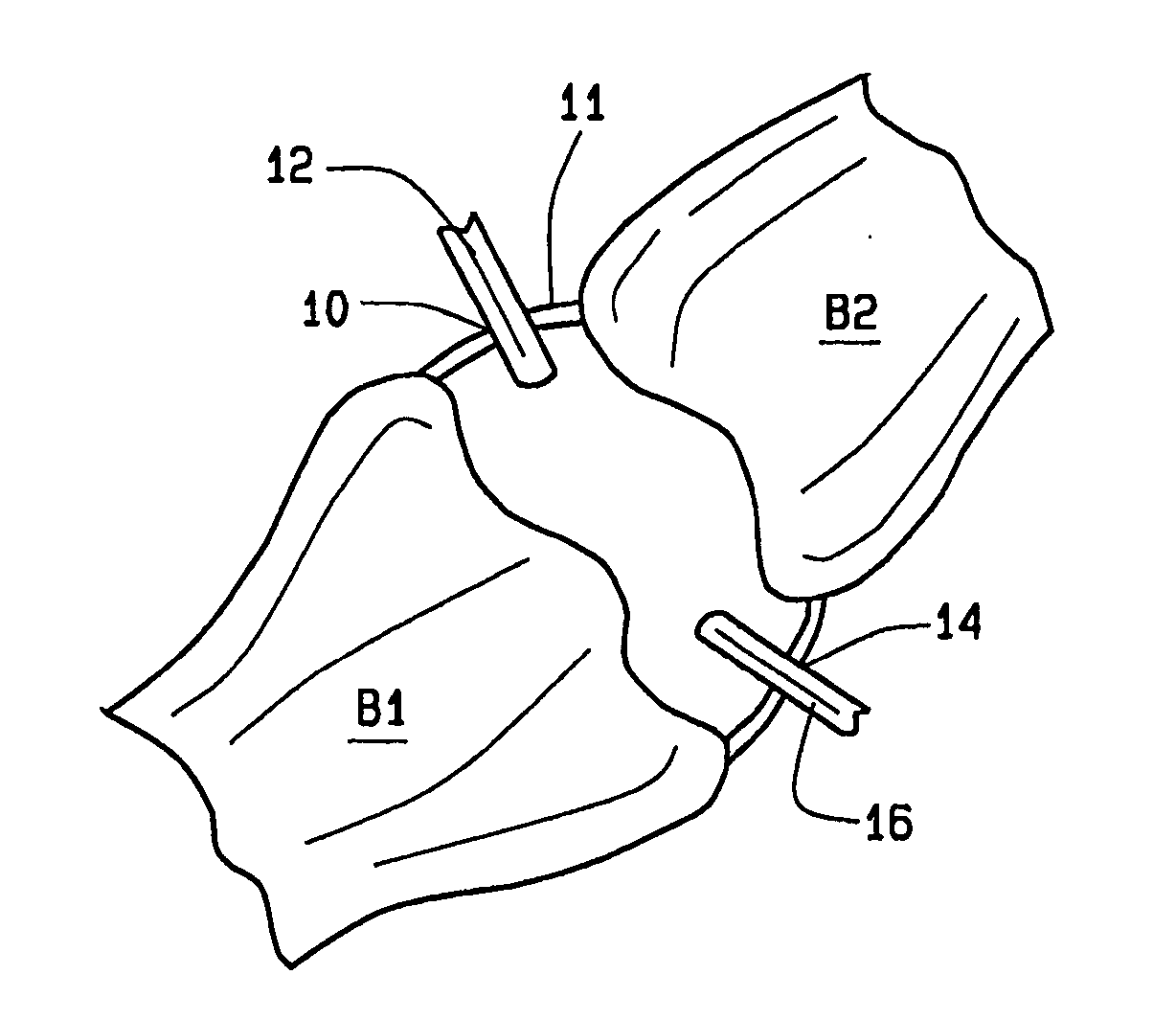
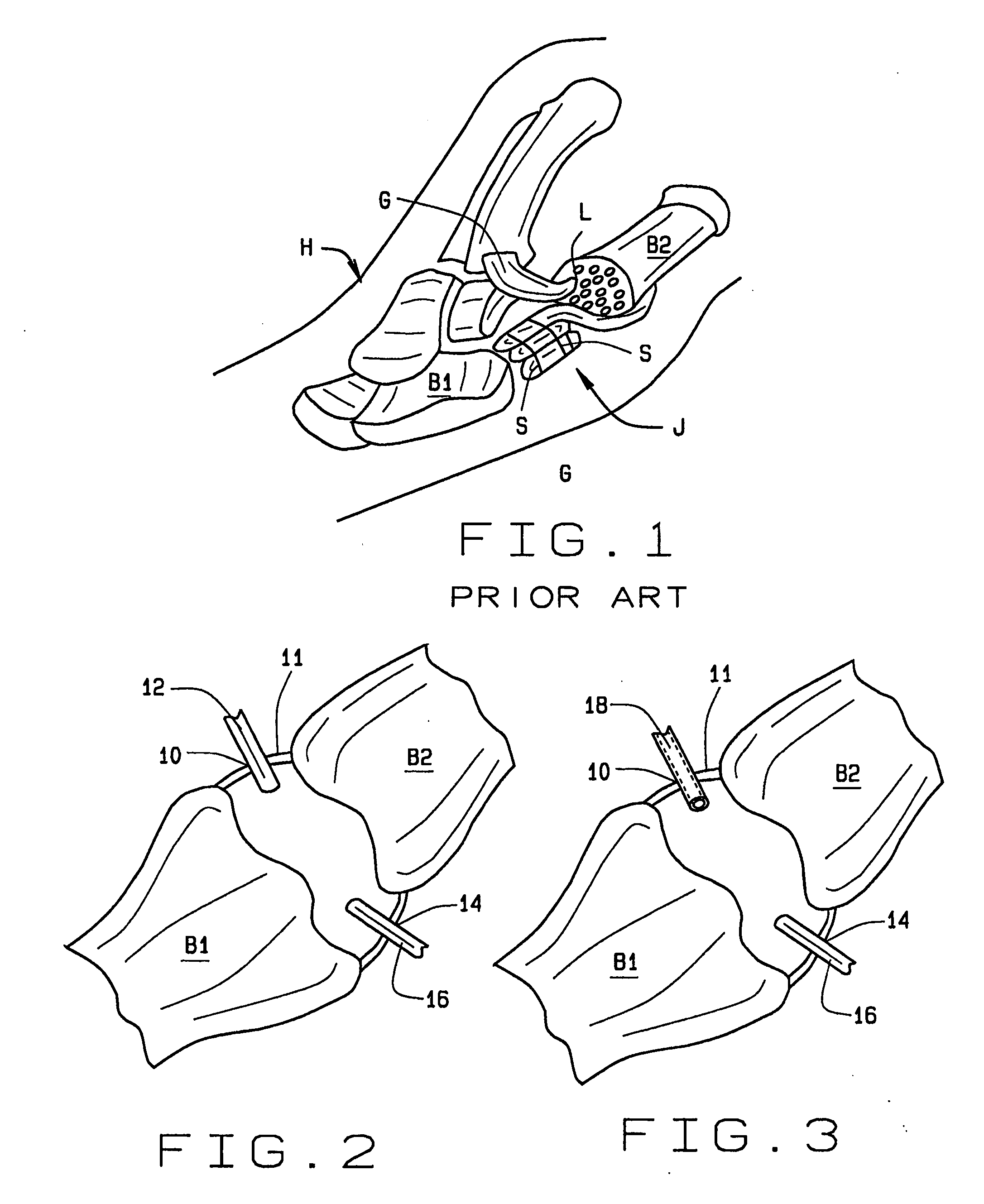
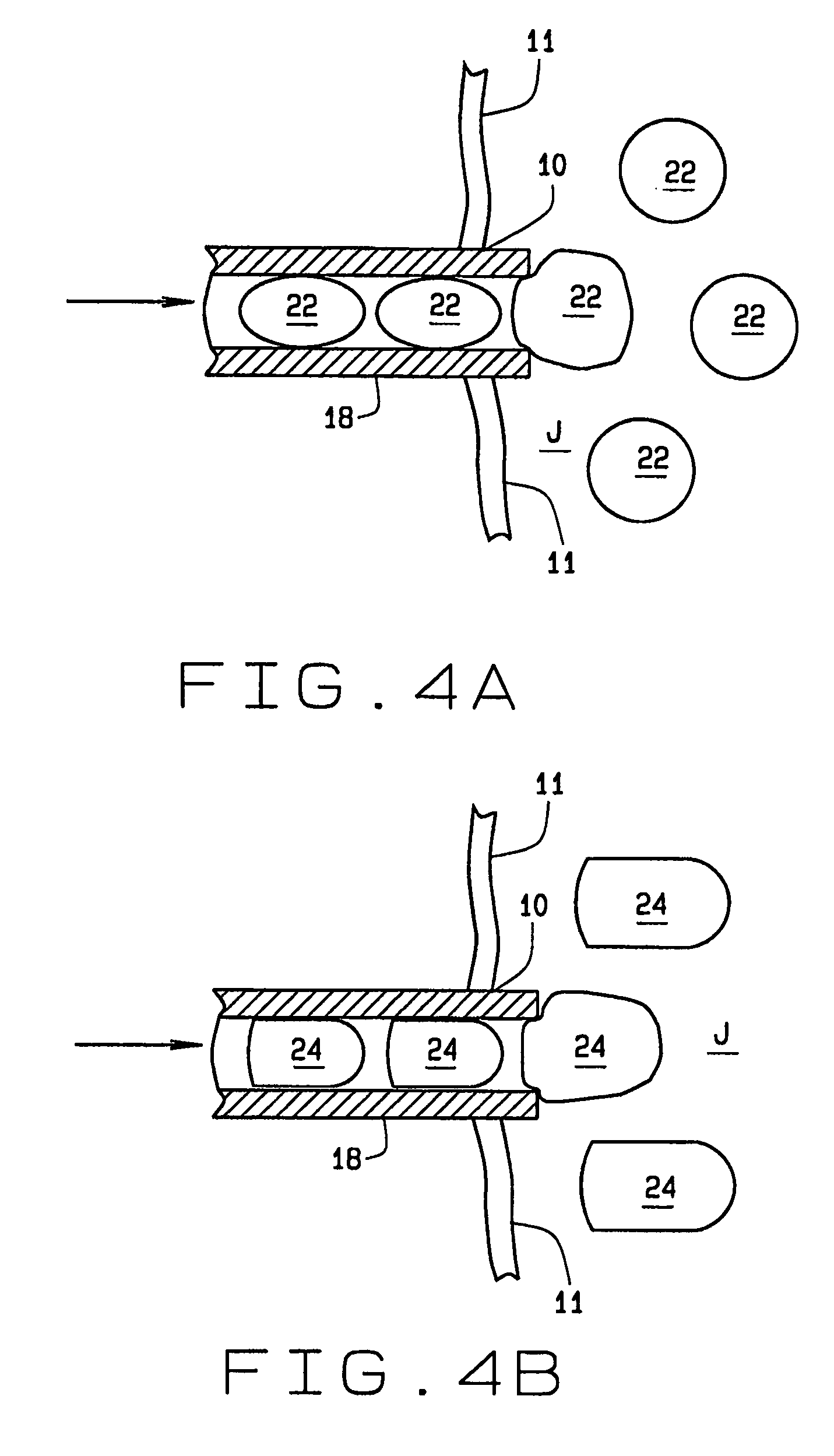
Arthoscopic arthroplasty procedure for the repair or reconstruction of arthritic joints
An arthoscopic arthroplasty procedure for the repair or reconstruction of an arthritic joint (J) in a person's hand or foot. A first opening hole (10) is made in a joint membrane (11), as is a second opening (14) for insertion of an arthoscope (16) into the joint to view the joint. A probe (12) is inserted into the joint to scrape away and remove arthritic material and diseased bone. A canula (18) is then inserted into the one opening for graft material, in the form of compressed pellets (22, 24), to be delivered to the joint. As the pellets exit the canula, they expand to a size which prevents their subsequent withdrawal from the joint through either hole. With the arthoscope, the surgeon is able to see that all the arthritic and diseased material is first removed from the joint, that the appropriate amount of graft material is then inserted into the joint to provide the cushioning required between the bones.
Owner:CHOW JAMES C Y
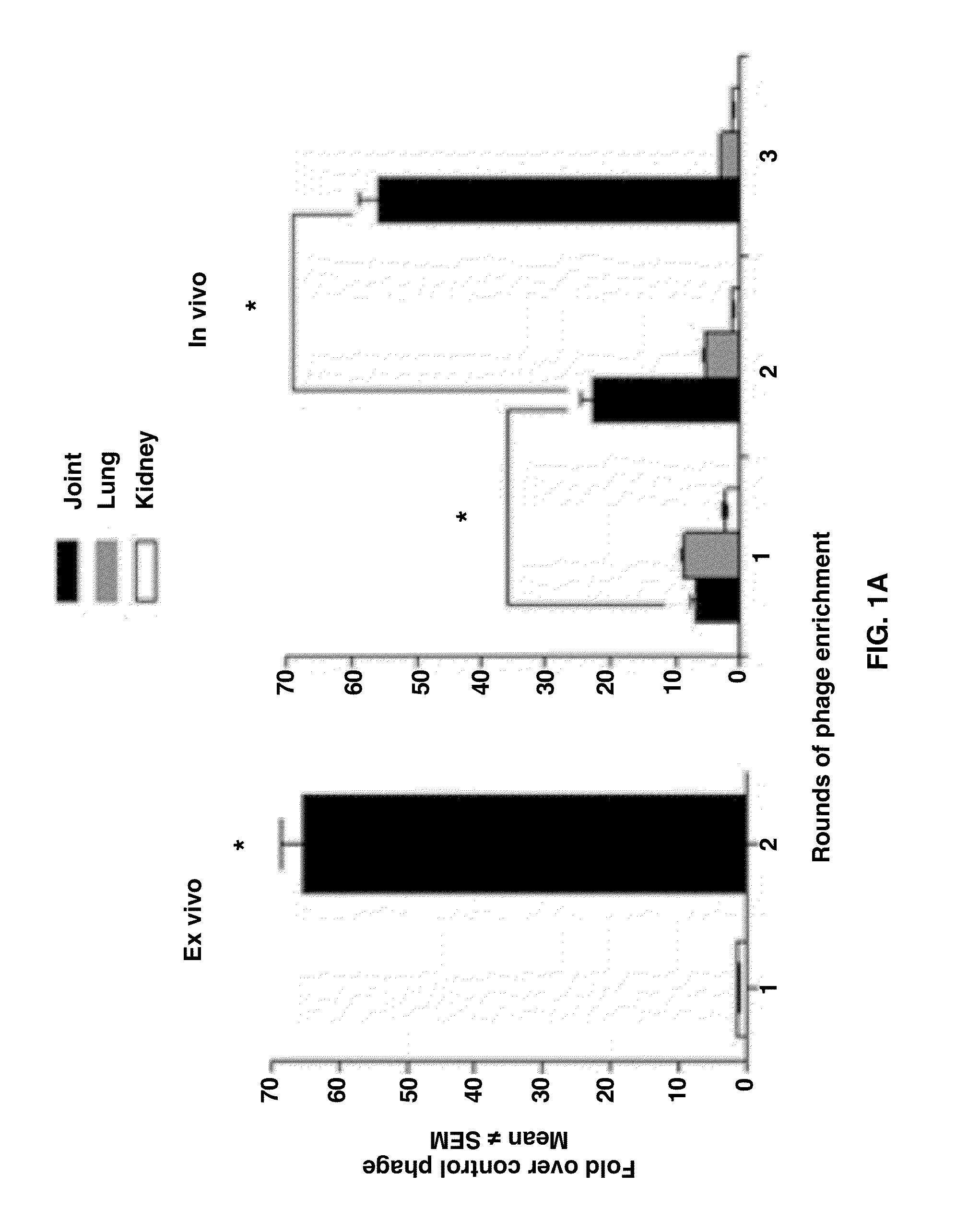
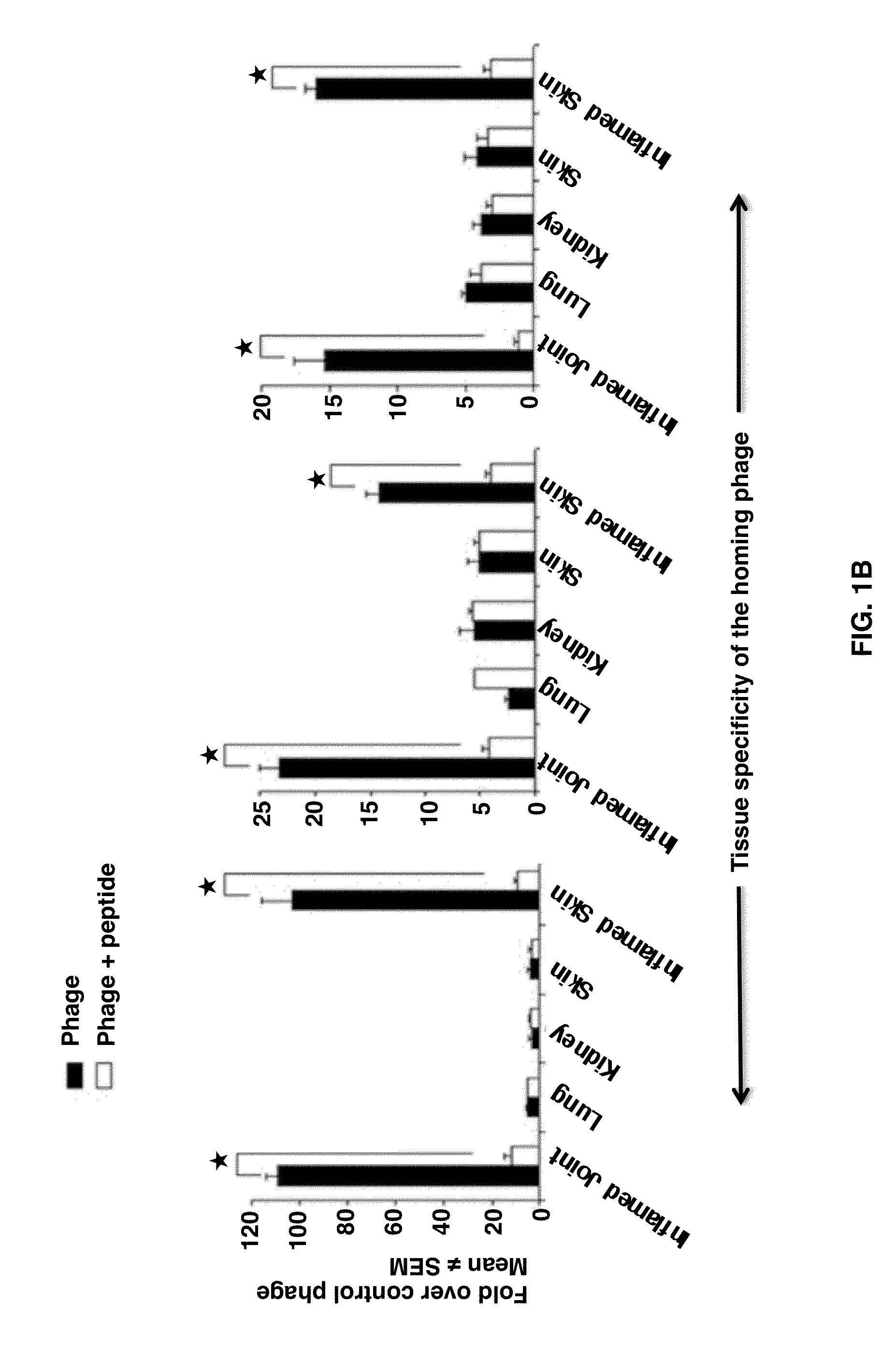
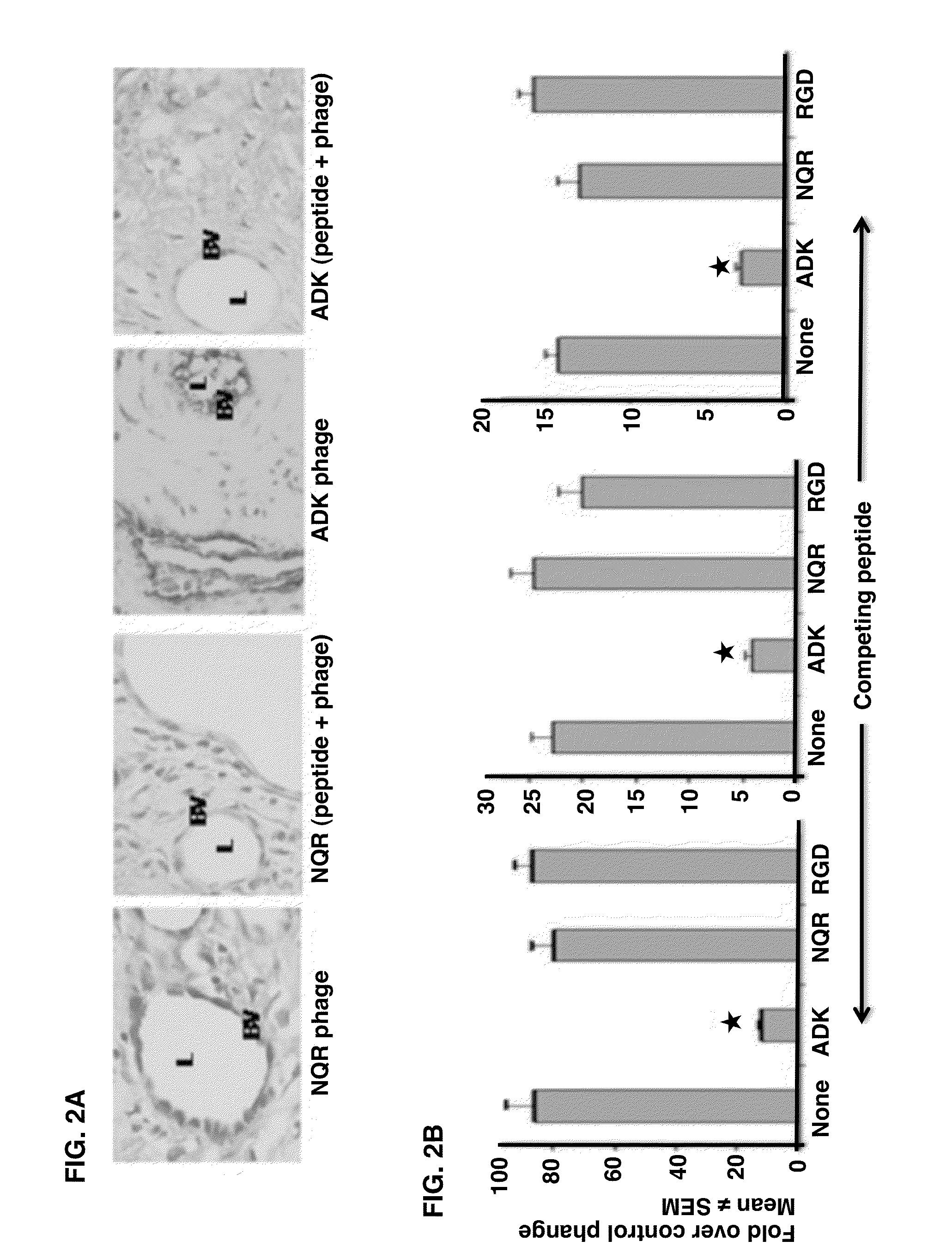
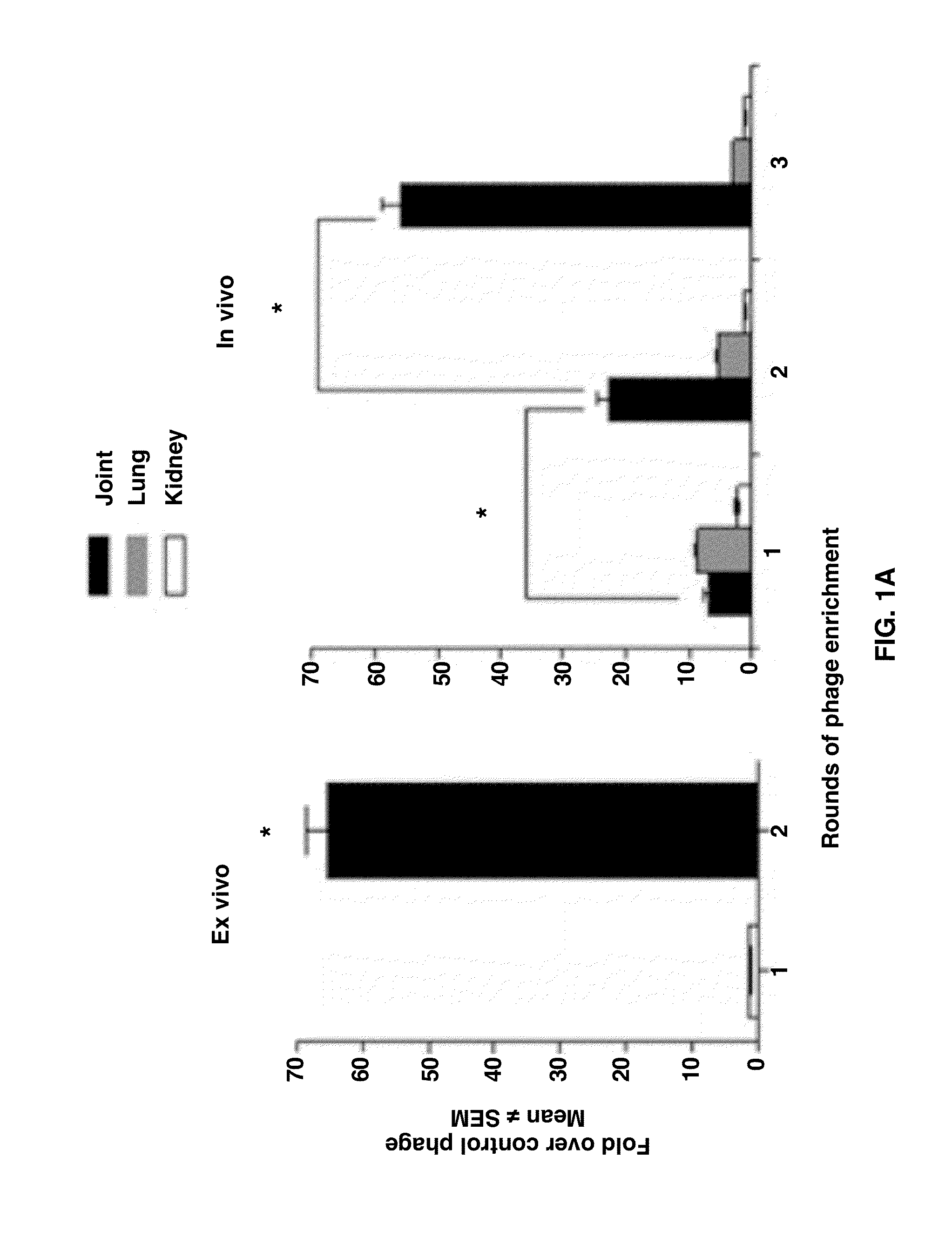
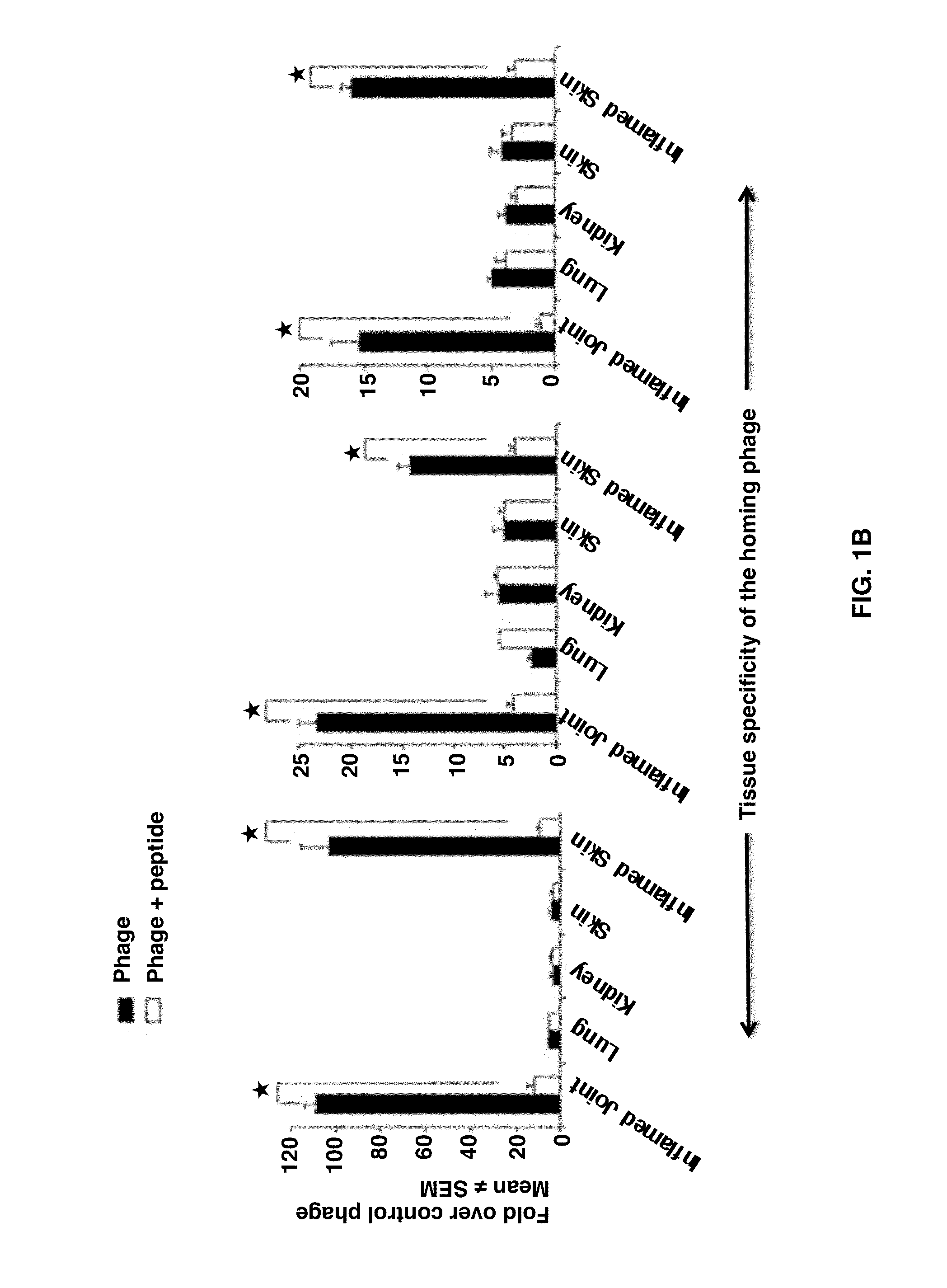
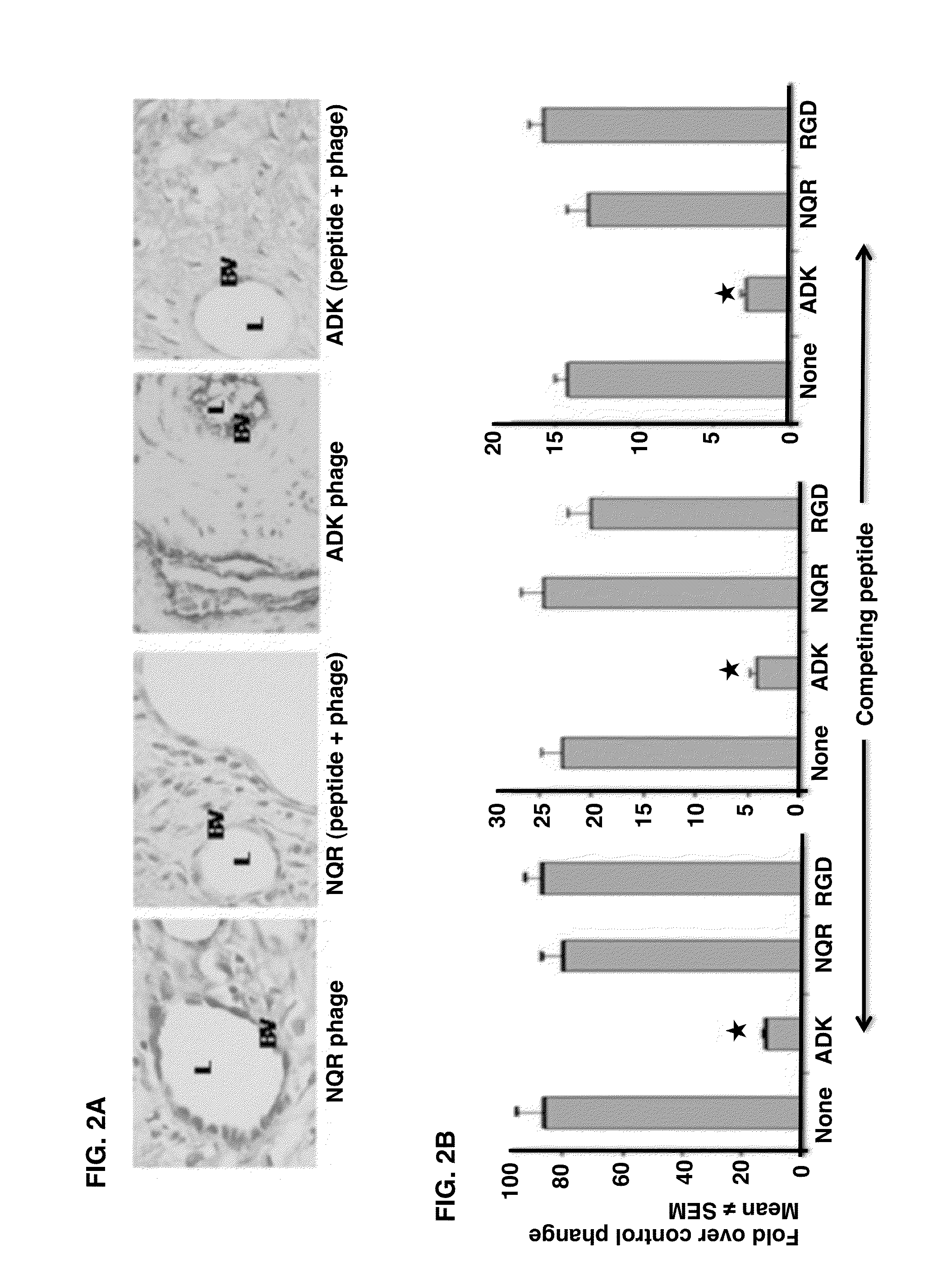
Joint-Homing Peptides and Uses Thereof
The present invention provides peptides that home to a joint of an animal, wherein said peptide comprises an amino acid motif selected from the group consisting of NQR and ADK. Also provided are methods of treating a subject having an arthritic joint, comprising the step of administering to said subject a pharmacologically effective dose of a composition provided herein.
Owner:UNIV OF MARYLAND BALTIMORE
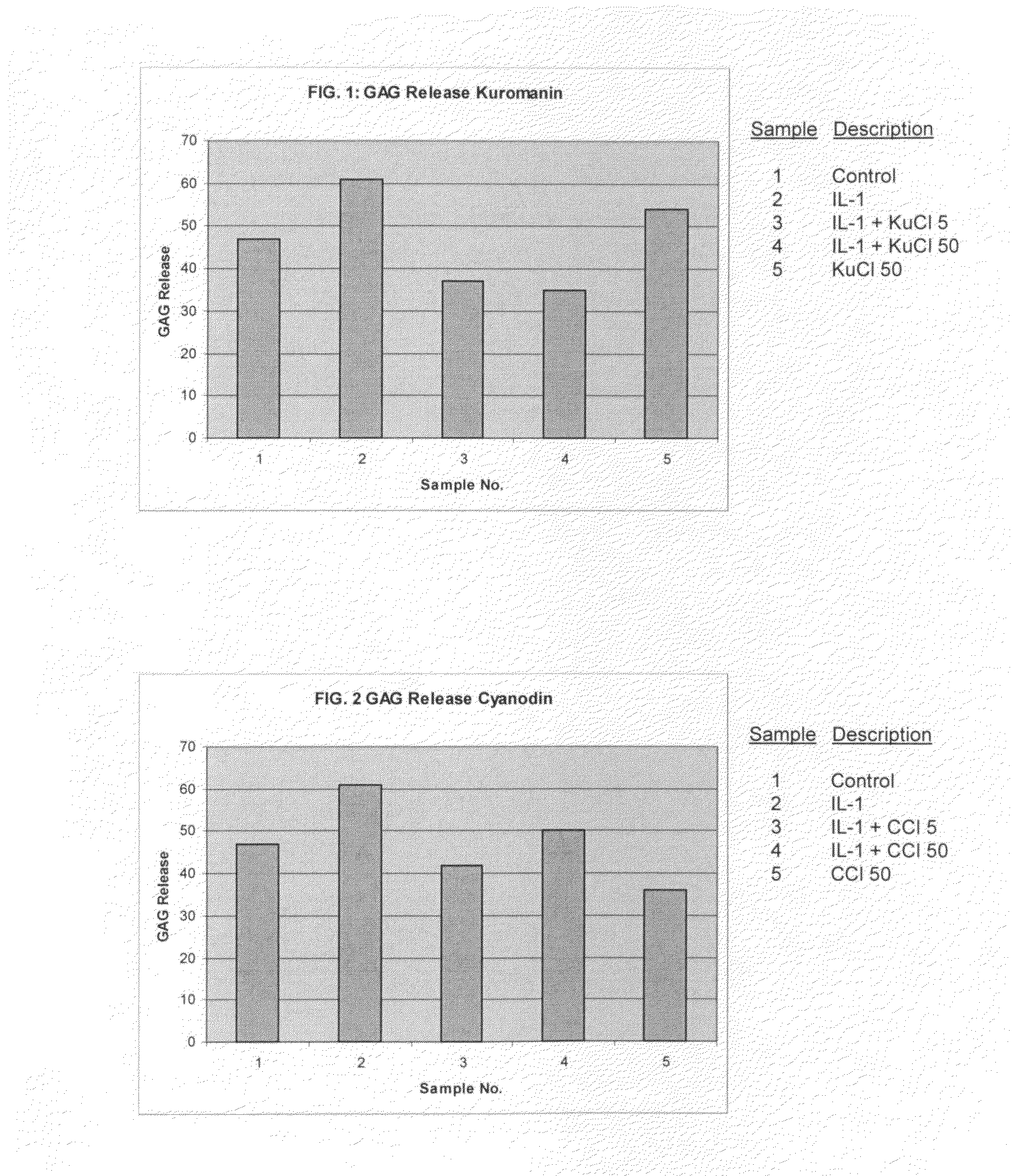
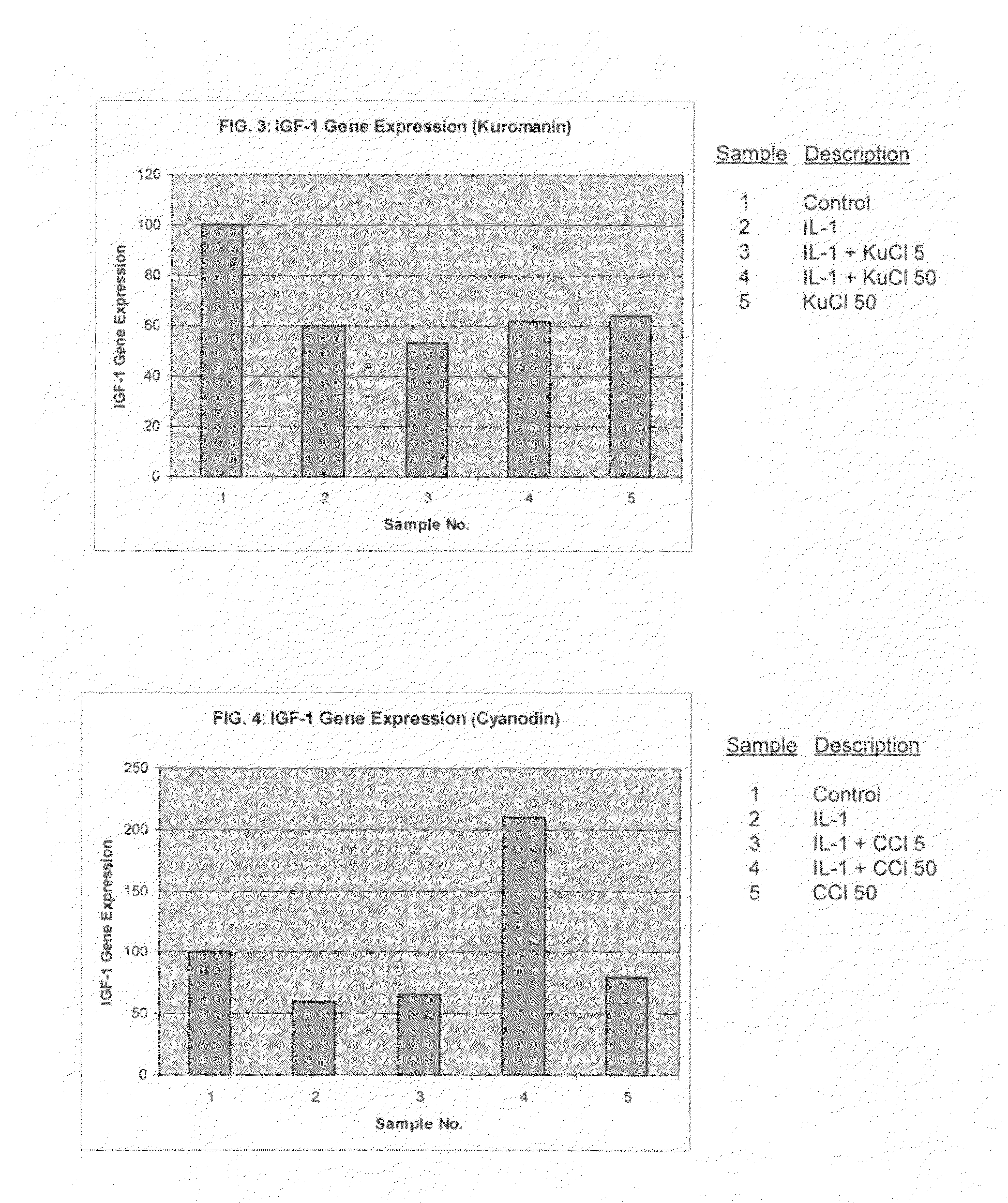
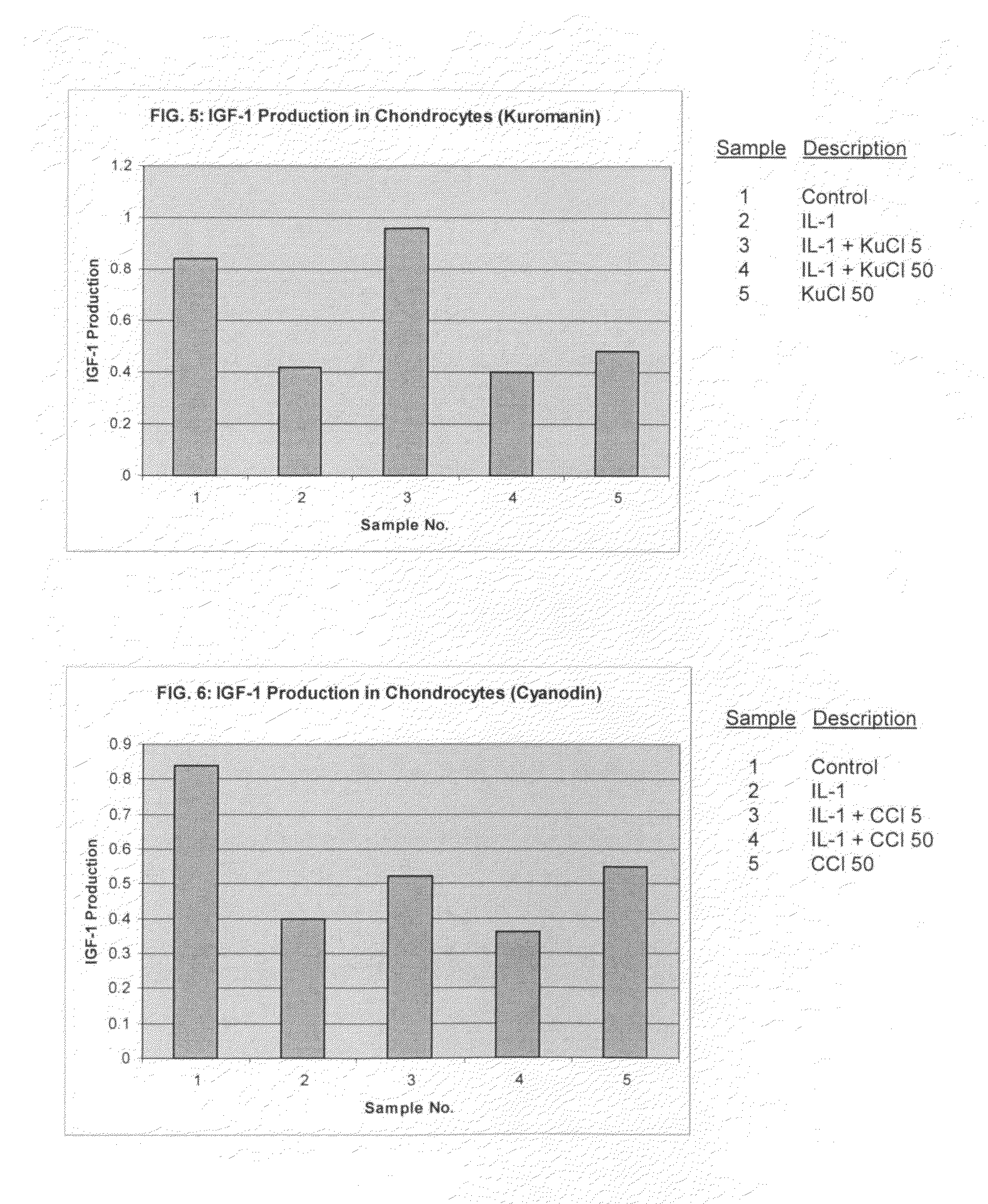
Compositions including anthocyanin or anthocyanidin for the prevention or treatment of articular cartilage-associated conditions
ActiveUS20100196331A1Prevent degradationEasy to produceBiocidePeptide/protein ingredientsArthritic jointSurgery
Methods of treating an arthritic joint of a subject, including administering a pharmaceutical composition by injection into the arthritic joint, wherein the composition includes an anthocyanin or anthocyanidin, glucose, and a pharmaceutically acceptable carrier.
Owner:JOHNSON LANNY L
Compound combined preparation for preventing osteoarticular pain and preparation method thereof
ActiveCN101822684AGood curative or preventive effectGood disintegration performancePeptide/protein ingredientsAntipyreticPharmaceutical preservativesPhosphate
The invention discloses a compound combined preparation for preventing osteoarticular pain and a preparation method thereof; the preparation comprises an active component glucosamine or combinations of more than one or two of the glucosamine and other active components chondroitin sulfate, dimethyl sulfoxide and collagen protein, and the glucosamine is glucosamine sulfate, hydrochloride, phosphate or the mixture thereof; and auxiliary materials comprise excipient, disintegration agent, lubricant, adhesive, stabilizer and wetting agent. The invention has good curing and preventing functions to injury in arthritis and joint movement, the prepared tablets have the advantages of good disintegration performance, high dissolution speed and quick effect, the used amount of the raw materials and all the auxiliary materials reaches the best ratio, the product stability is improved, and the absorption utilization rate is high.
Owner:JIANGSU ALAND NOURISHMENT
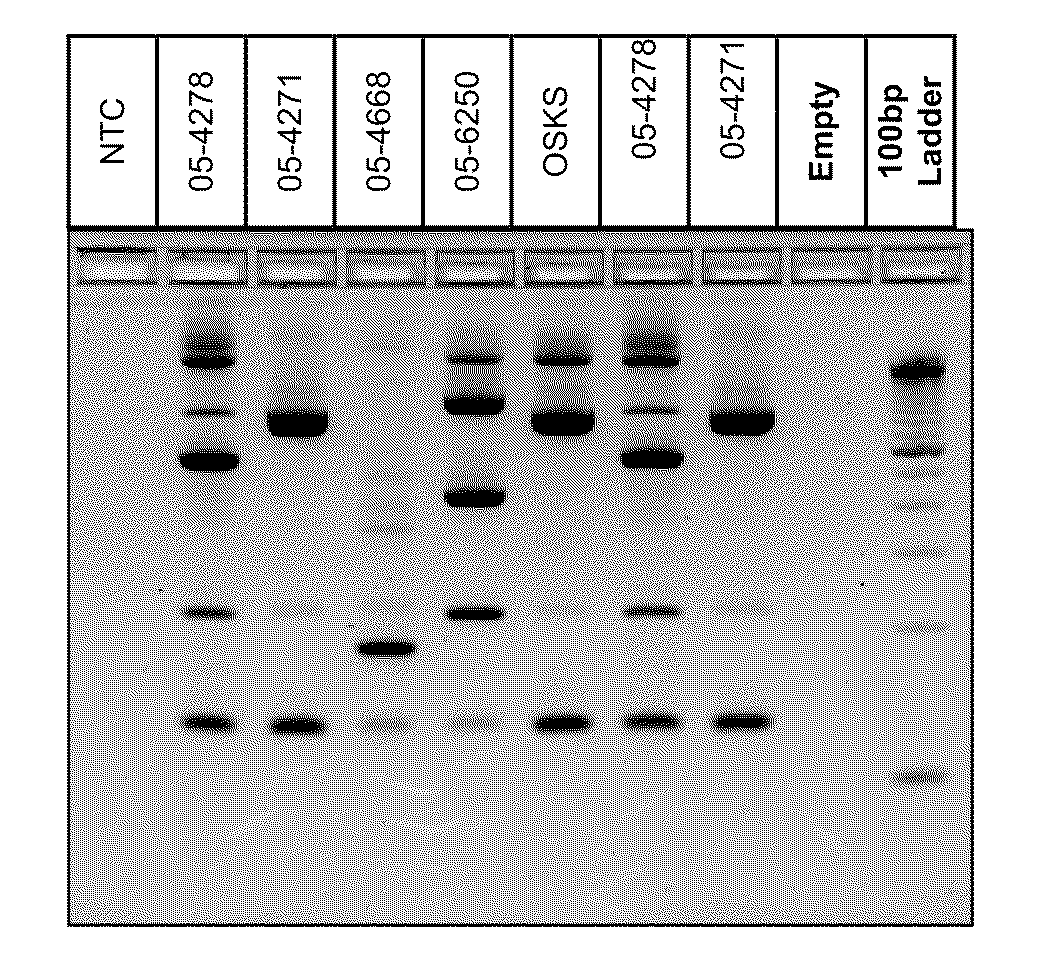
Pcr-based genotyping
InactiveUS20110059437A1Improve analysisHigh sample throughputSugar derivativesMicrobiological testing/measurementInsertion sequenceBiology
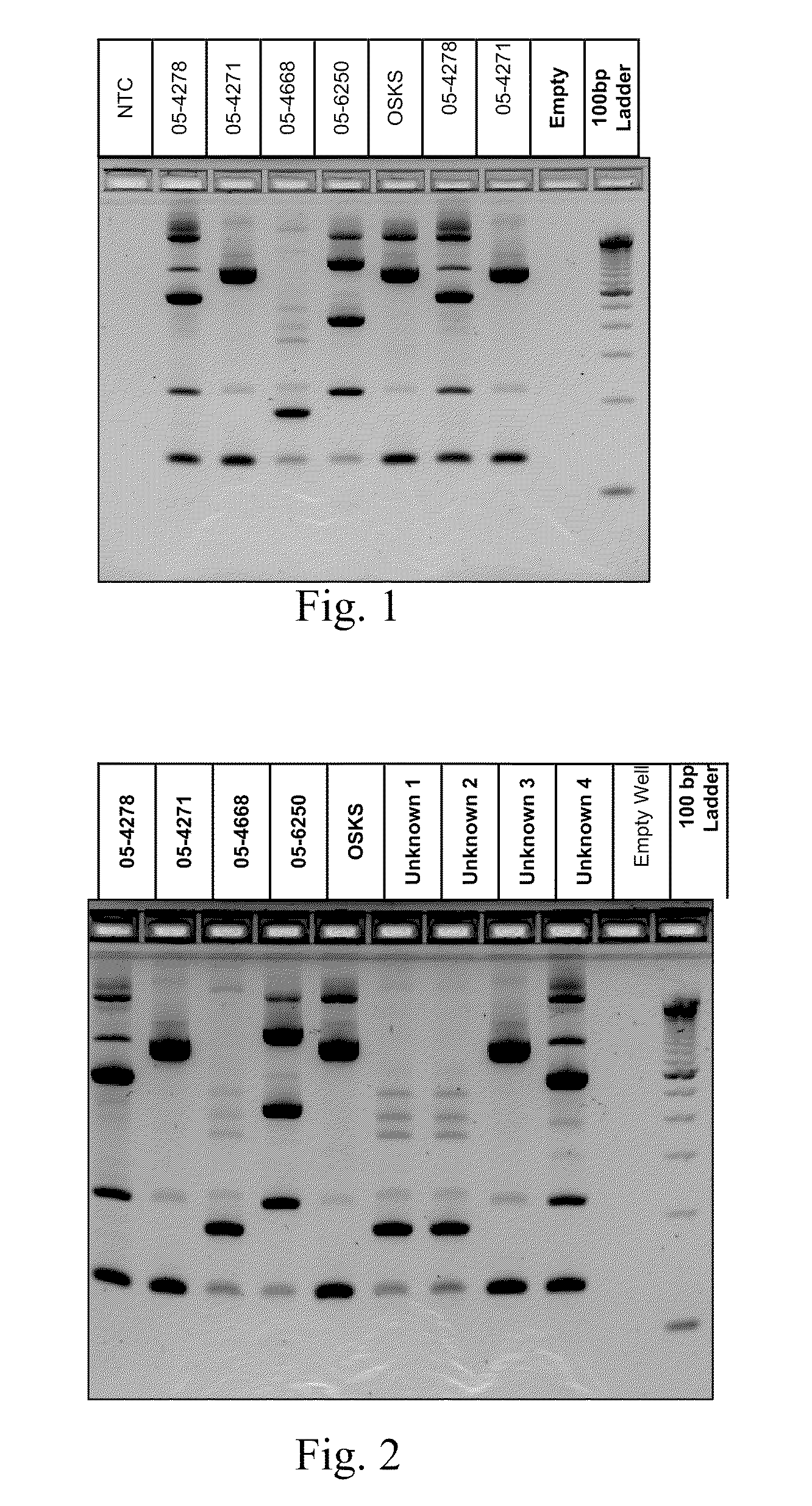
A Mycoplasma bovis PCR-based genotyping method was developed that exploits the proximity of insertion sequences (IS) within the genome by using outward facing primers that selectively amplify sequences between IS elements. The method was applied to 16 field isolates of M. bovis, originating from pneumonic lung or arthritic joints, collected from the United States (Iowa or Kansas) between 2004 and 2005. The genomic fingerprints generated 14 distinct amplification profiles consisting of 4-8 fragments ranging in size from 200-3000 bp. Three isolates presented identical patterns and were isolated from two calves (one calf with pneumonic lung and the other with both pneumonic lung and arthritic joint) from a single farm during an outbreak and probably represent multiple infections with the same genotype. To demonstrate the stability of IS markers for molecular fingerprinting, 3 of the 16 field isolates were subjected to high number passage which resulted in patterns identical to the initial isolates. The results of these studies demonstrate the method can be used for simple and rapid molecular fingerprinting and differentiating M. bovis isolates with extension to epidemiology.
Owner:BOEHRINGER LNGELHEIM VETMEDICA GMBH
Compositions including anthocyanin or anthocyanidin for the prevention or treatment of articular cartilage-associated conditions
ActiveUS8263069B2Prevent degradationEasy to produceBiocidePeptide/protein ingredientsArthritic jointSurgery
Methods of treating an arthritic joint of a subject, including administering a pharmaceutical composition by injection into the arthritic joint, wherein the composition includes an anthocyanin or anthocyanidin, glucose, and a pharmaceutically acceptable carrier.
Owner:JOHNSON LANNY L
Novel peptide complexes
The invention relates to novel self-assembling peptide complexes comprising a charged peptide and a polysaccharide, methods of producing them and uses therefor. The novel self-assembling peptide complexes of the present invention have particular utility in the restoration of biomechanical or biochemical function of a variety of biological tissues, for example and without limitation, in degenerated spinal discs, osteoarthritic joints, damaged cartilage, meniscus, ligaments, tendons, dental, ophthalmic and cardiovascular and blood vessel tissues. The invention provides inter alia methods of repairing and or restoring biomechanical or biochemical function of biological tissues and scaffolds for the support of cell growth.
Owner:UNIV OF LEEDS
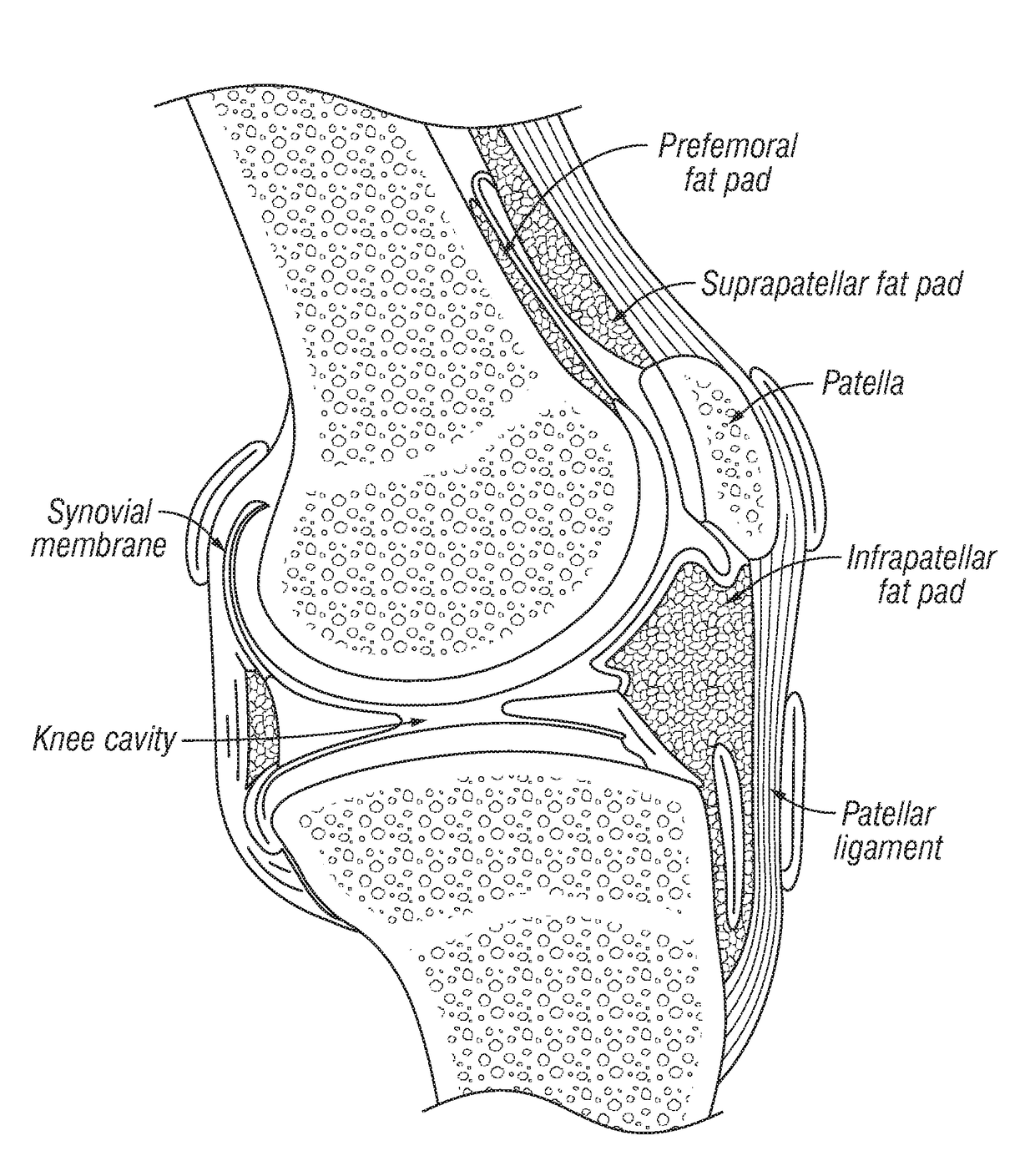
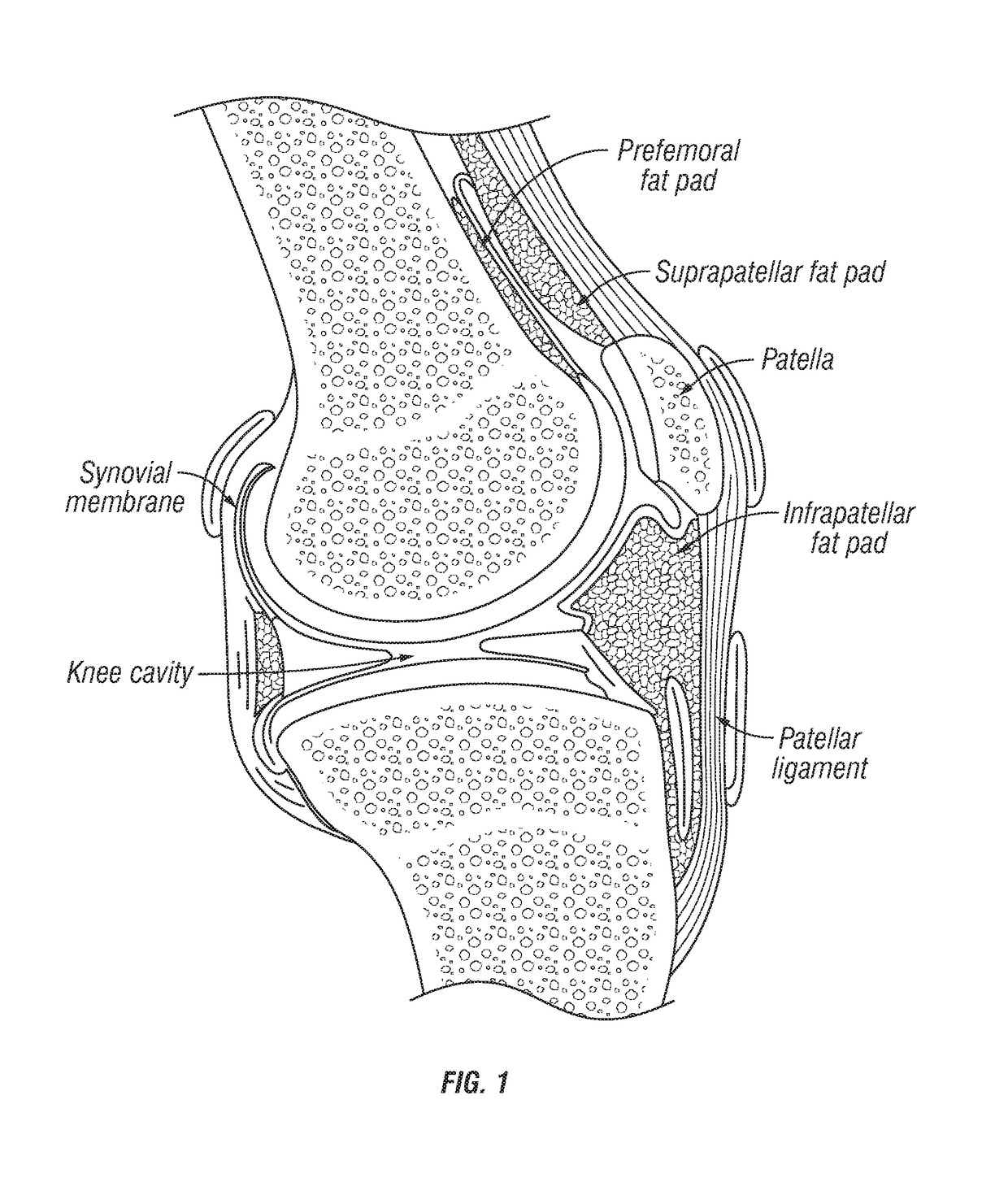
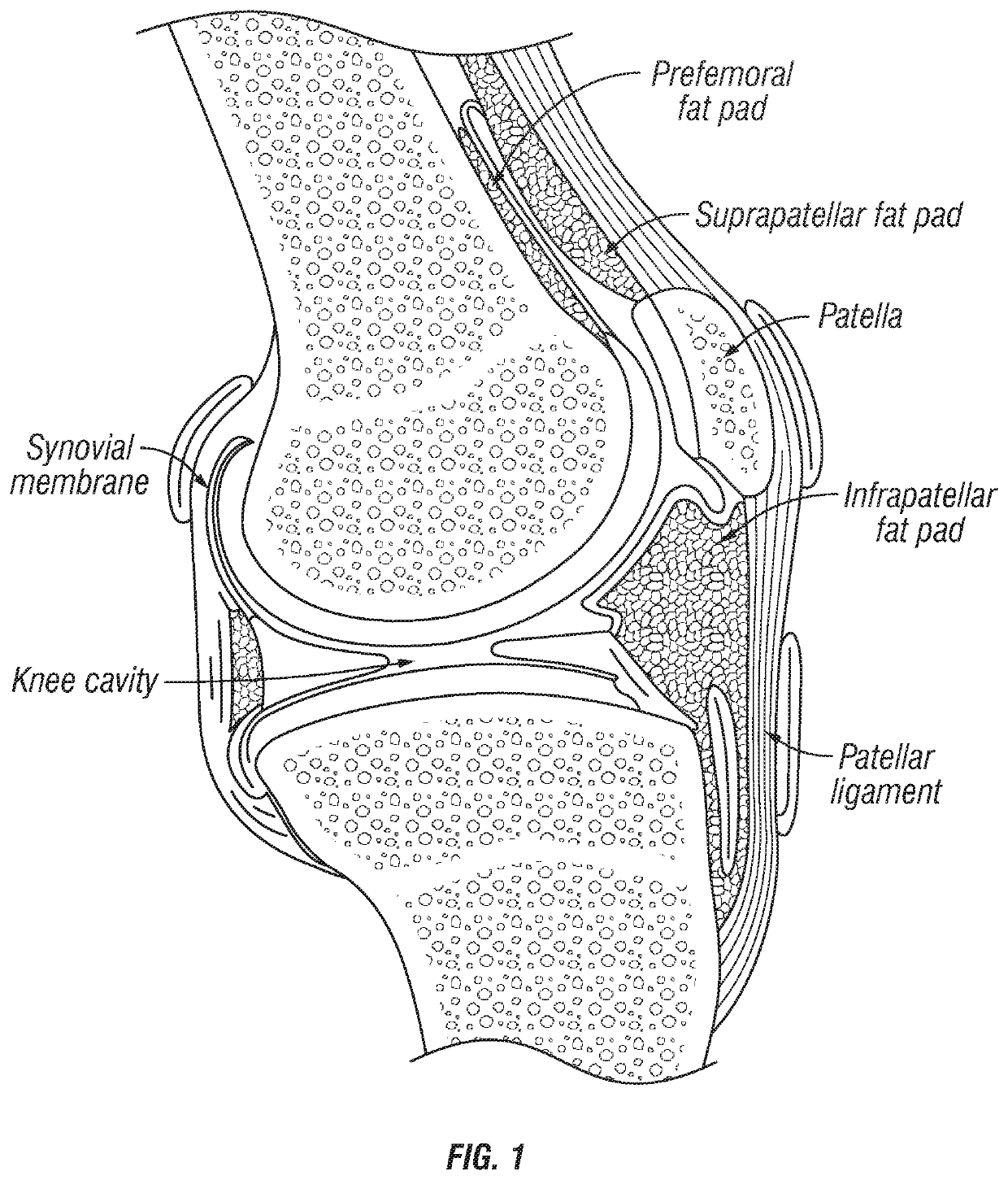
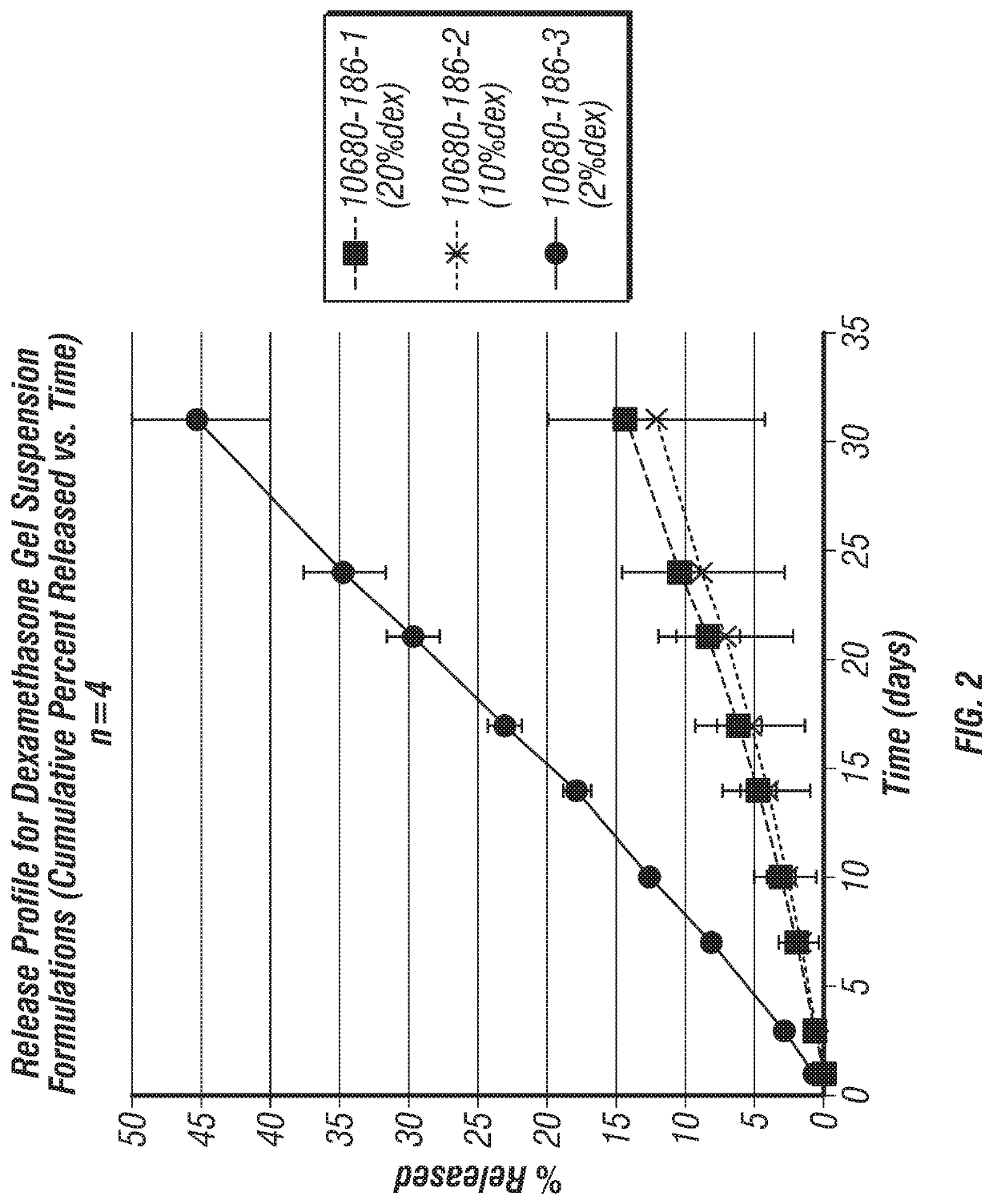
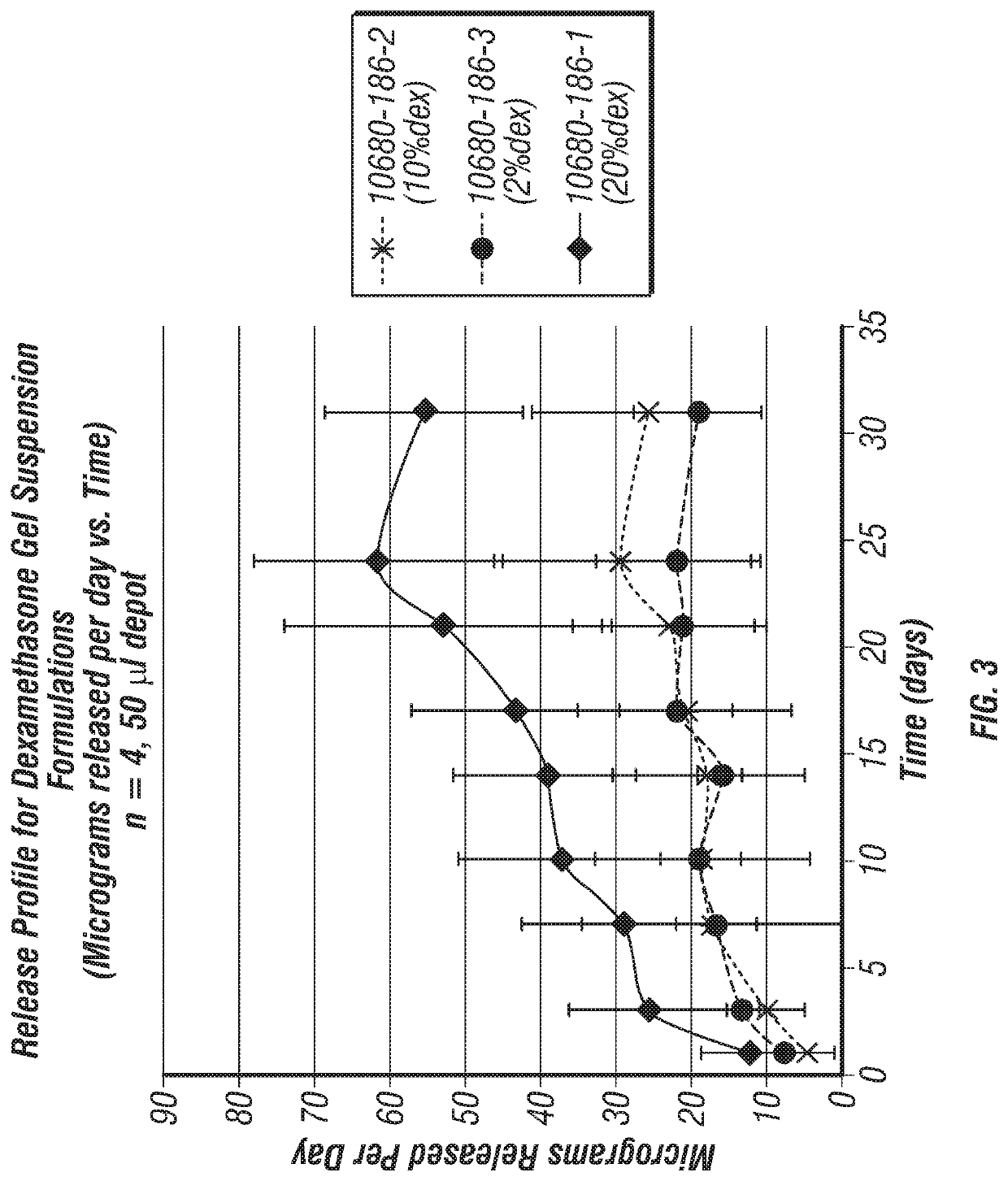
Joint fat pad formulations, and methods of use thereof
ActiveUS20170368236A1Reduced systemic exposureImprove dose accuracyOrganic active ingredientsNervous disorderDiseaseWhole body
The present invention relates to formulations for administration to a joint fat pad of a subject, and to methods of treating joint pain, inflammation or disease. The disclosed formulations are intended for local administration to the joint fat pad to provide sustained release of a therapeutic agent to the joint cavity and surrounding tissues. The joint may be an arthritic joint, an injured joint or a surgically replaced joint. The therapeutic agent may be an analgesic agent, an anti-inflammatory agent or an immunosuppressive agent. A single administration of the formulation to the joint fat pad delivers a therapeutically effective amount of the therapeutic agent with reduced systemic exposure relative to a single systemic or a single intra-articular administration of a therapeutic dose of an identical therapeutic agent.
Owner:ALLERGAN INC
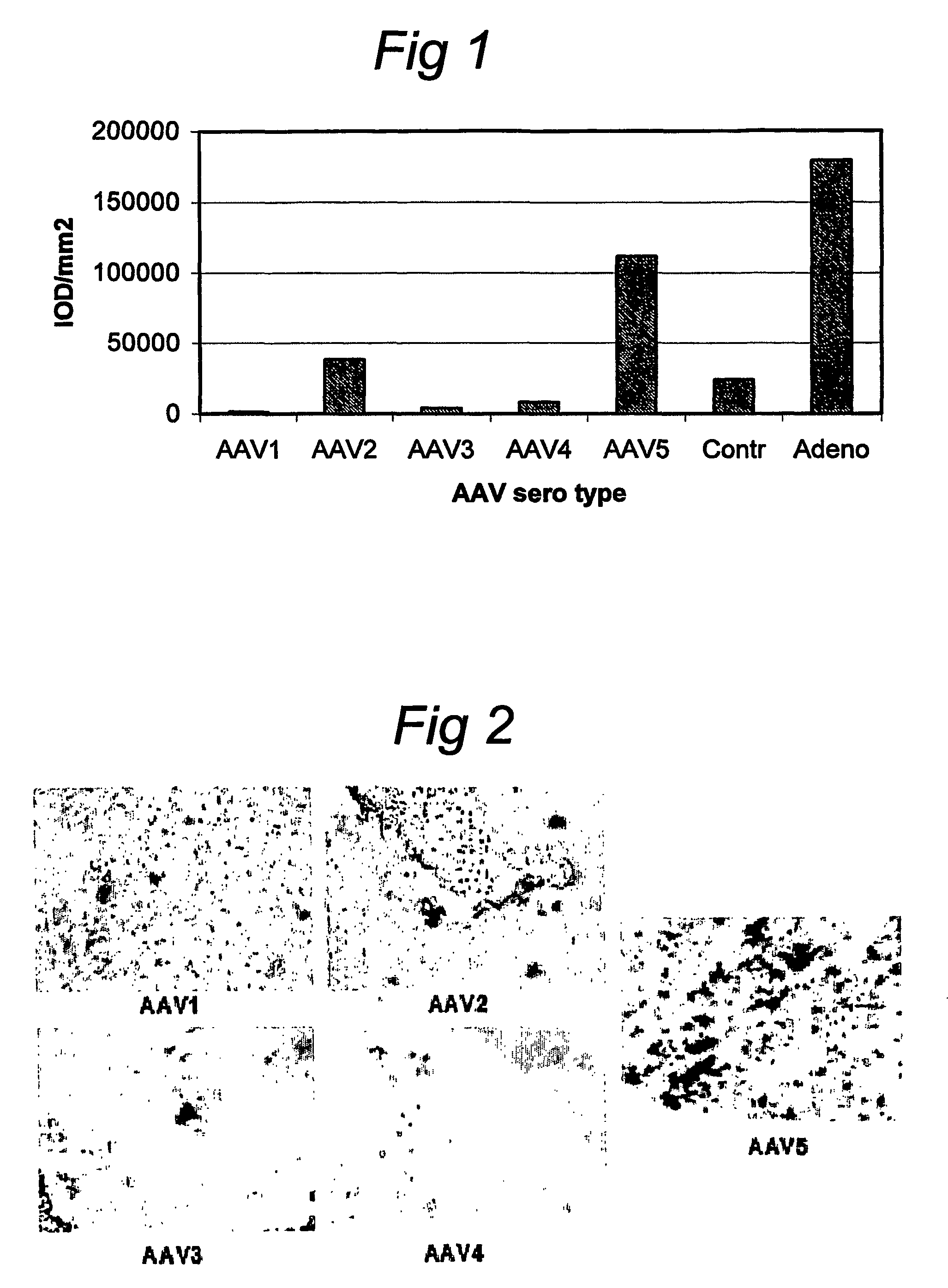
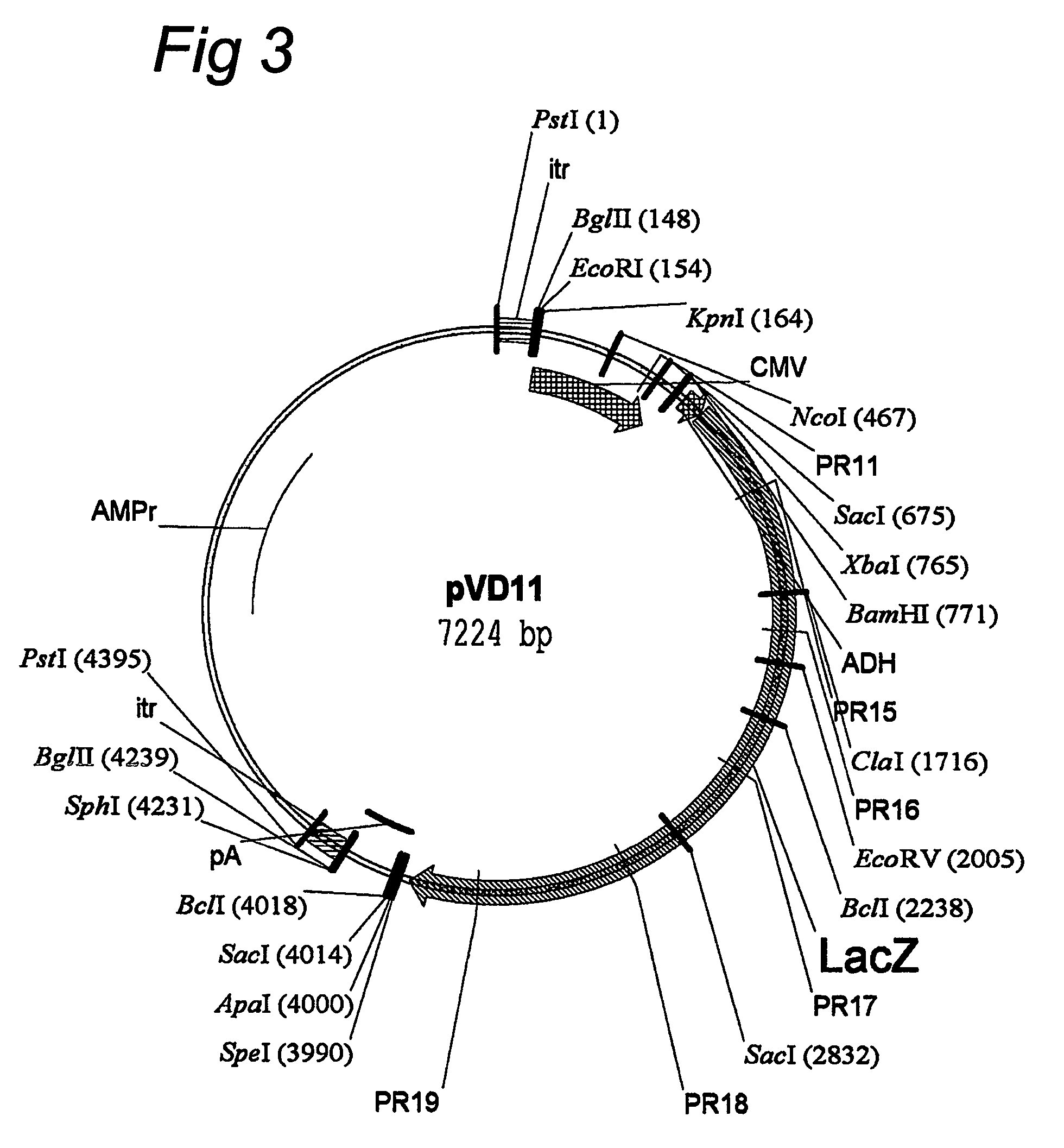
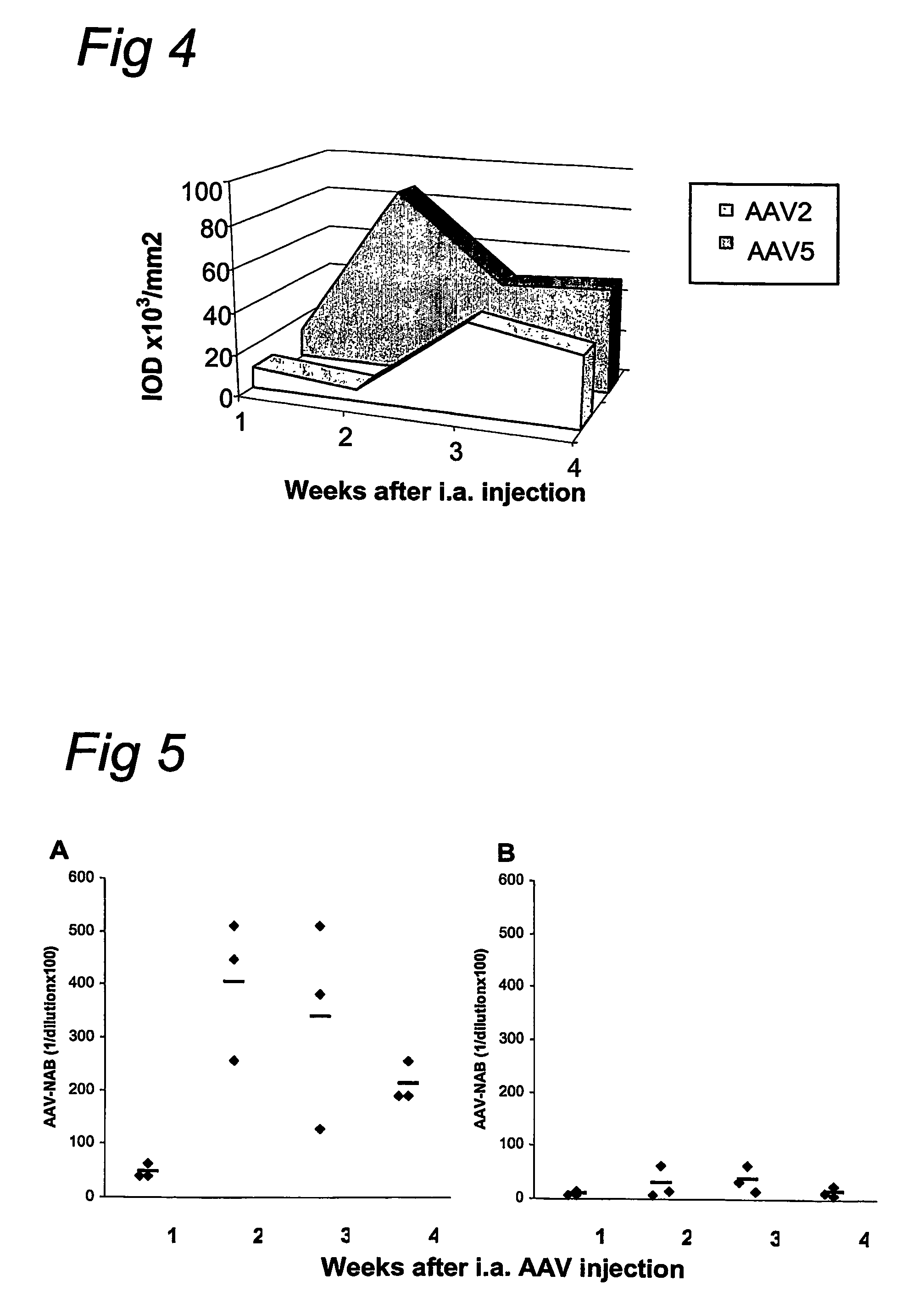
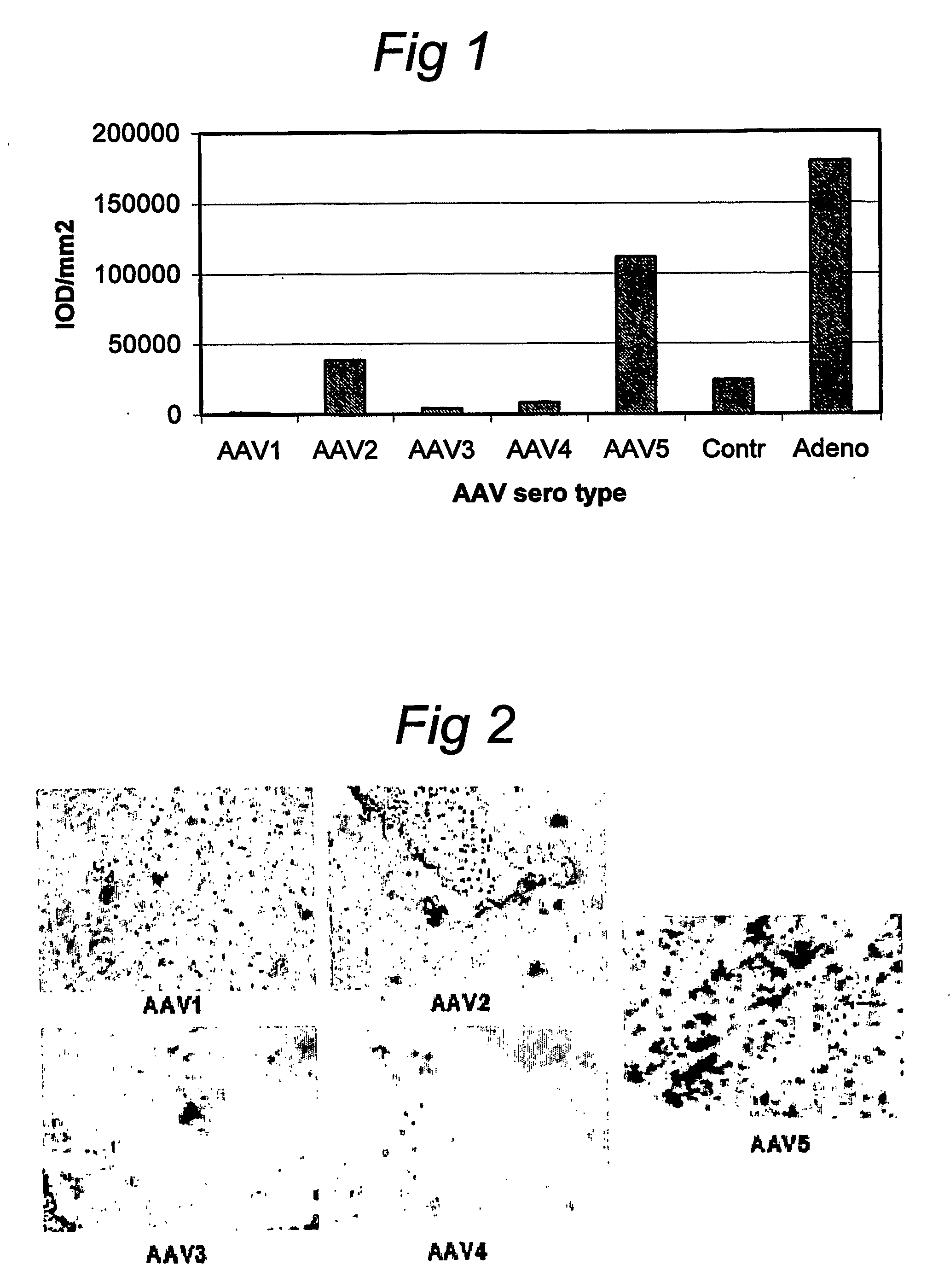
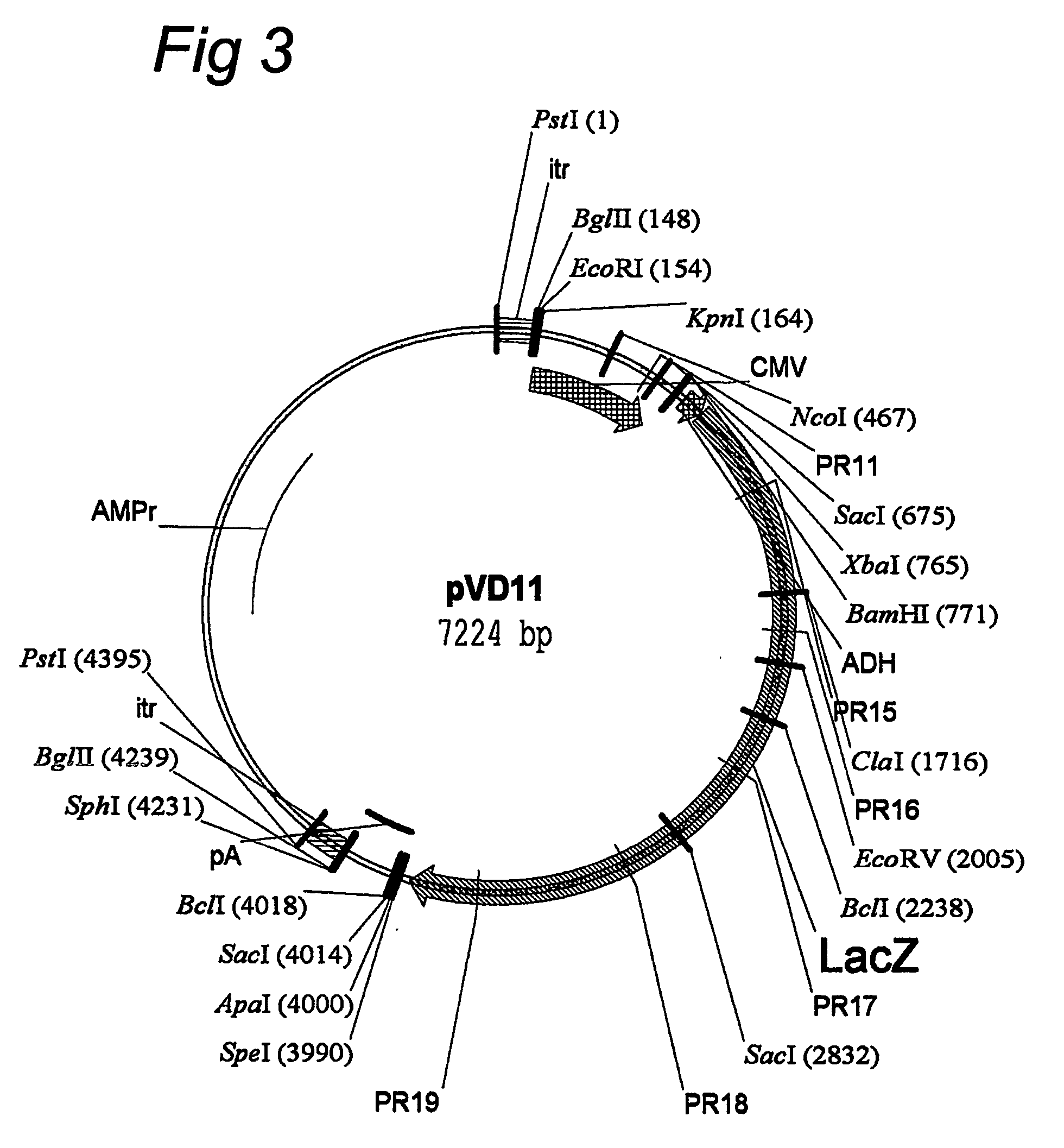
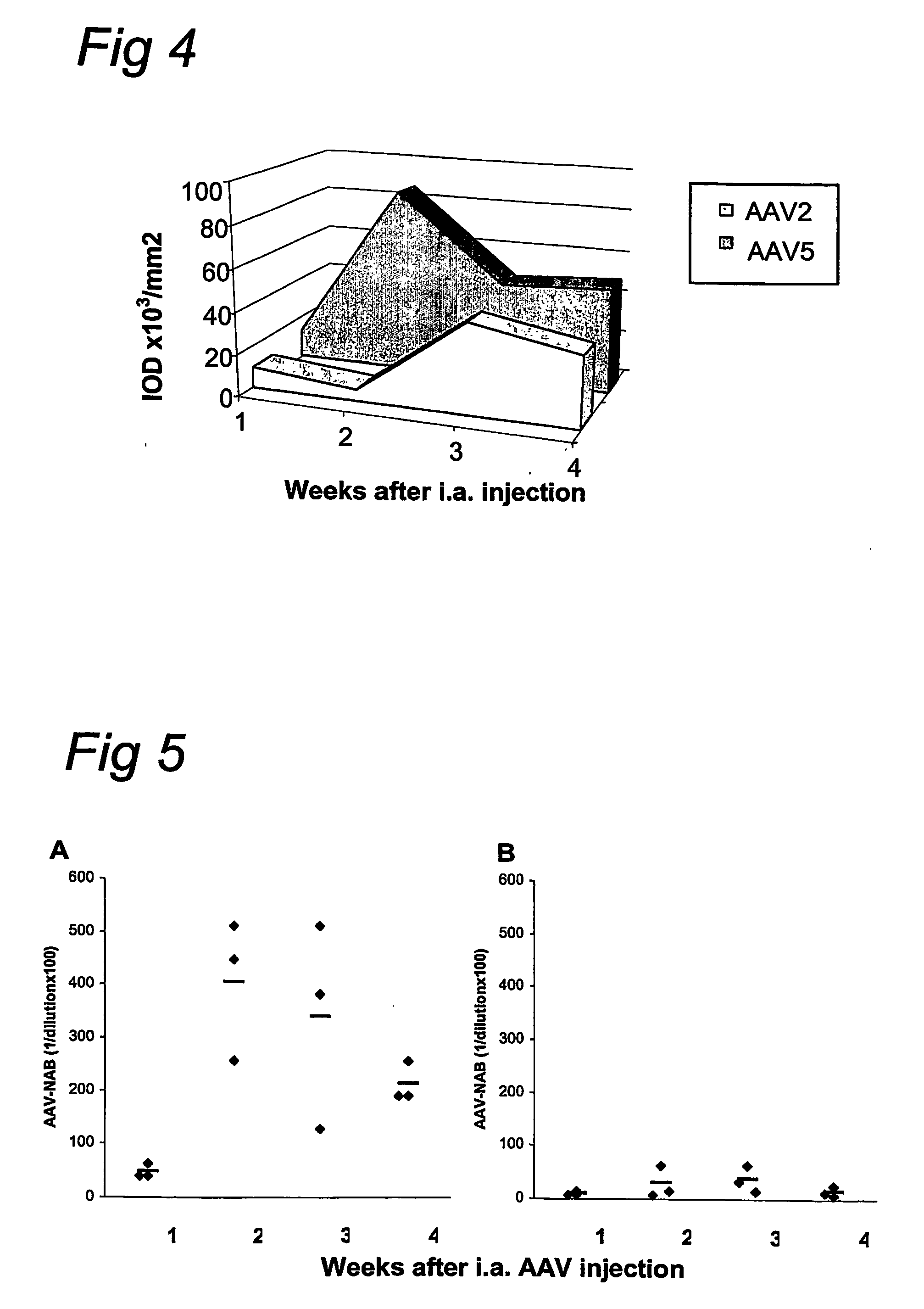
AAV vectors for in vivo gene therapy of rheumatoid arthritis
The present invention relates to the field of adeno-associated virus (AAV) based gene therapy, in particular in vivo gene therapy, of rheumatoid arthritis (RA). The invention provides recombinant AAV virions being highly efficient in delivering genes encoding therapeutic proteins to the arthritic joints, and method for using such virions in in vivo gene therapy.
Owner:ACADEMISCH MEDISCH CENT BIJ DE UNIV VAN +1
Application of akebia saponin D to preparation of medicines for promoting osteoarthritic cartilage repair
InactiveCN108159063APromote differentiationAvoid damageOrganic active ingredientsAntipyreticArticular painMedicine
The invention discloses the application of akebia saponin D to preparation of medicines for promoting osteoarthritic cartilage repair. Tests prove that the akebia saponin D can promote differentiationof mesenchymal stem cells into chondrocytes and relieve osteoarthritic articular cartilage damage.
Owner:刘丽宏
Joint-homing peptides and uses thereof
The present invention provides peptides that home to a joint of an animal, wherein said peptide comprises an amino acid motif selected from the group consisting of NQR and ADK. Also provided are methods of treating a subject having an arthritic joint, comprising the step of administering to said subject a pharmacologically effective dose of a composition provided herein.
Owner:UNIV OF MARYLAND BALTIMORE
Meniscus for joint reconstruction
The invention provides treatment methods of using meniscus to repair injured and / or arthritic joints, for example, small hand joints including but not limited to radiocarpal, metacarpophalangeal, and interphalangeal joints. The invention also provides various implants made of meniscus for injured and / or arthritic joints.
Owner:CEDARS SINAI MEDICAL CENT
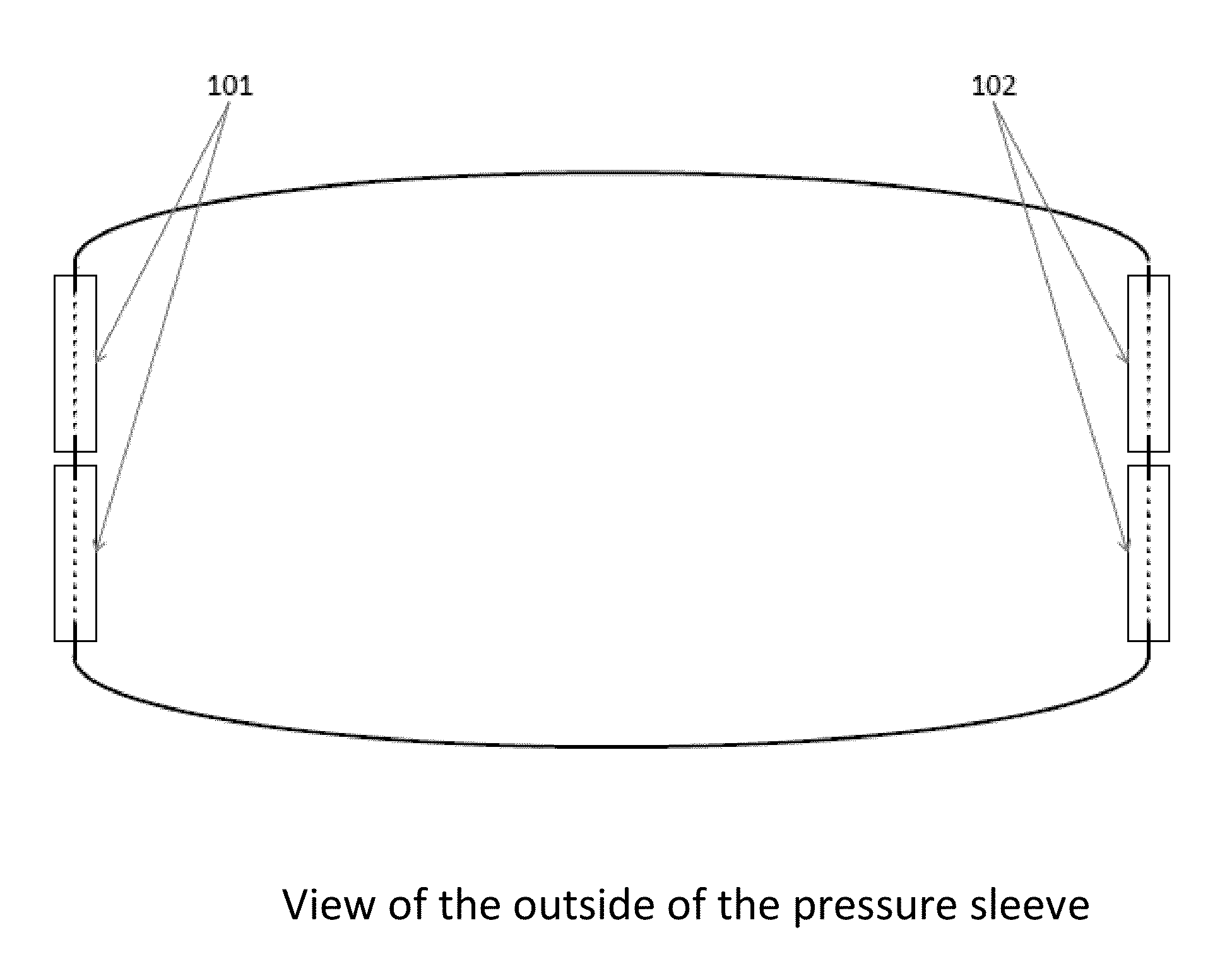
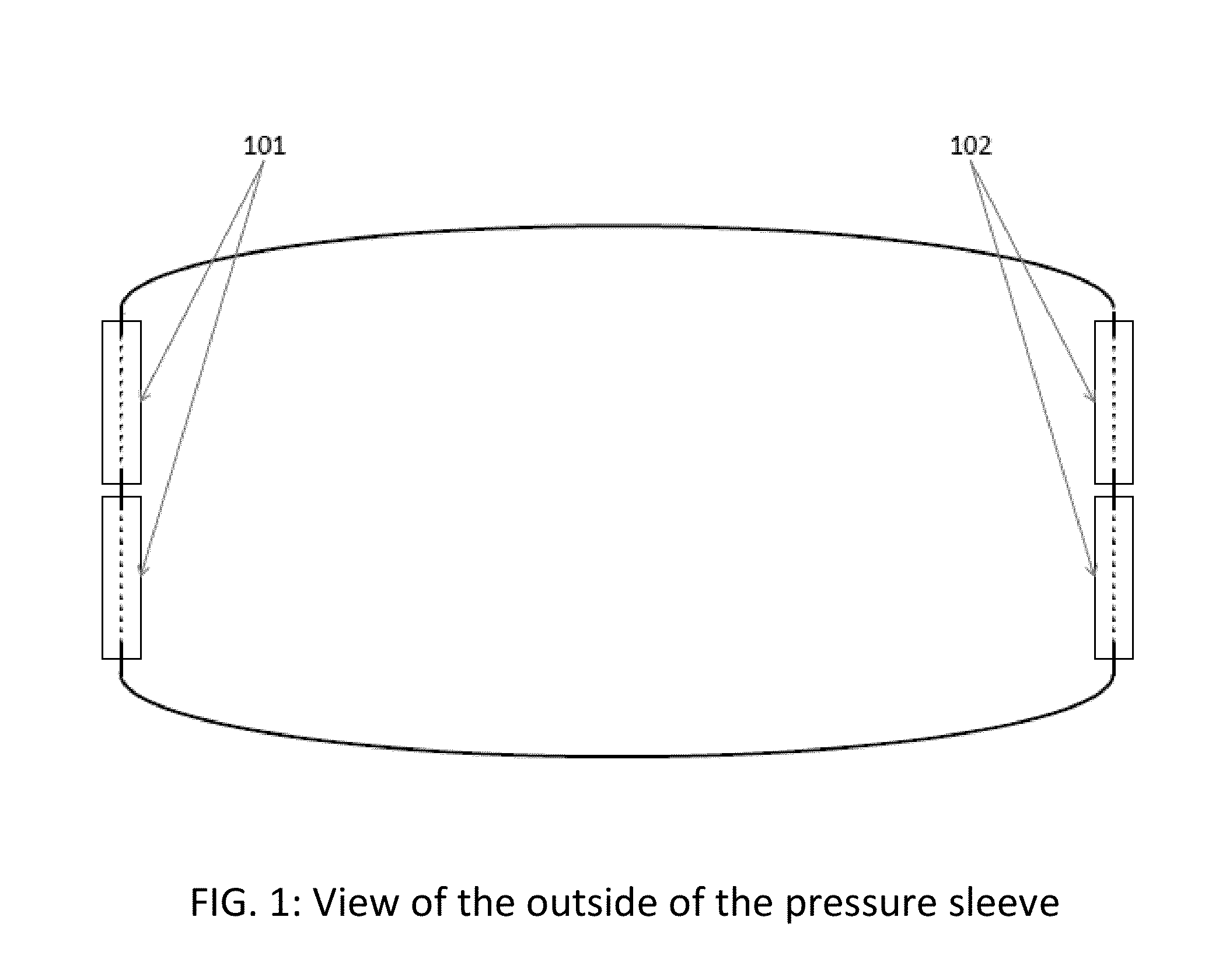
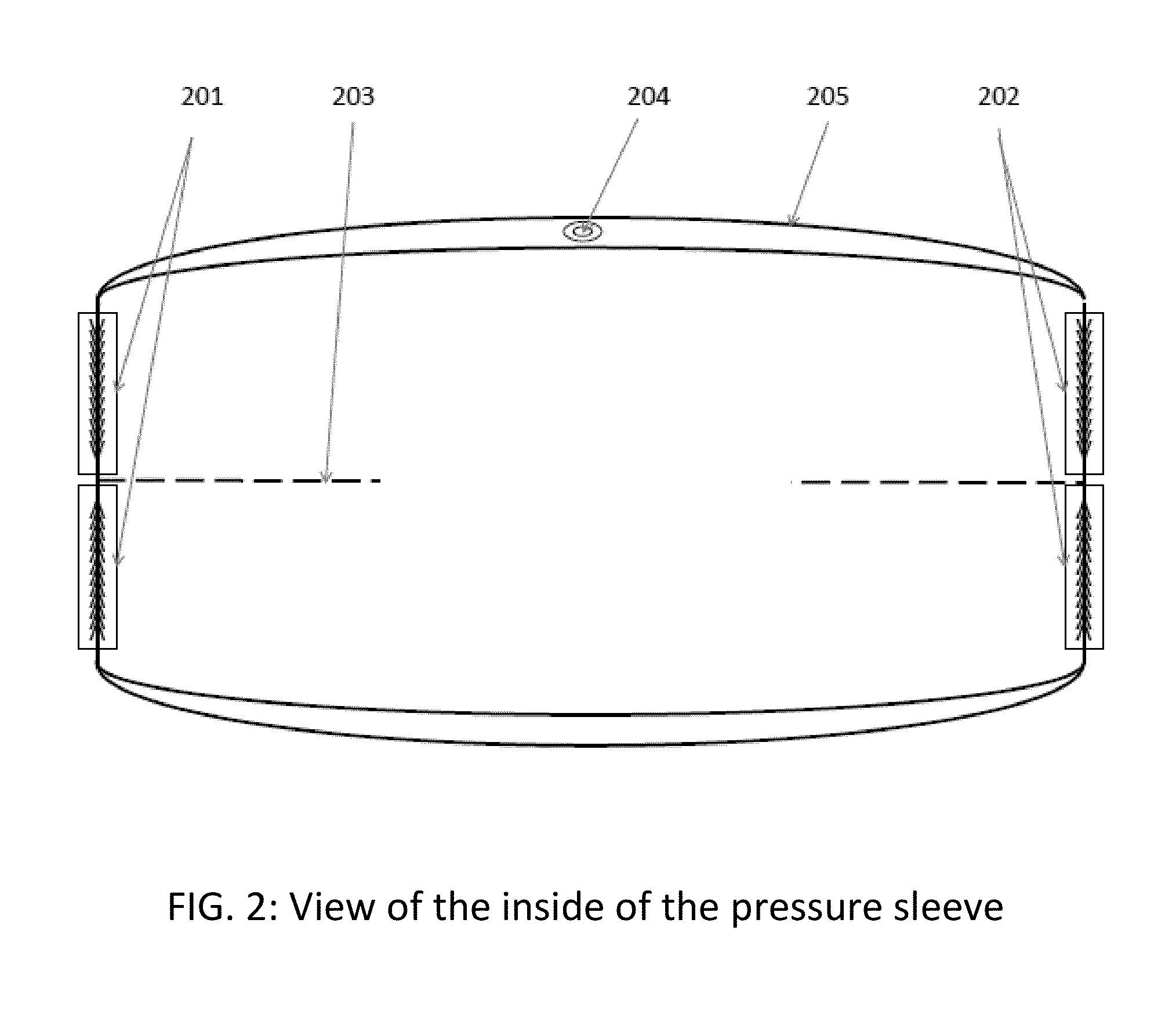
Apparatus and method for relieving weather related arthritic joint pain
InactiveUS20140378877A1Relief of arthritic joint painSerious painMassage combsPneumatic massagePain aggravatedArthritic joint
Apparatus and method for relief of arthritic joint pain aggravated due to change in weather, especially cold, rainy weather. The invention offers a practical method to address this pain while allowing user mobility and without being seen with overt medical contraptions. The invention consists of a specialized pressure sleeve with air at a specified temperature and pressure, design to fit joints and specialized clothing to fasten the sleeves. The method describes the usage of the apparatus to achieve the said goal.
Owner:AGARWAL SAKSHI +1
AAV vectors for in vivo gene therapy of rheumatoid arthritis
The present invention relates to the field of adeno-associated virus (AAV) based gene therapy, in particular in vivo gene therapy, of rheumatoid arthritis (RA). The invention provides recombinant AAV virions being highly efficient in delivering genes encoding therapeutic proteins to the arthritic joints, and method for using such virions in in vivo gene therapy.
Owner:ACADEMISCH MEDISCH CENT BIJ DE UNIV VAN +1
Macromolecular Delivery Systems for Non-Invasive Imaging, Evaluation and Treatment of Arthritis and Other Inflammatory Diseases
This invention relates to biotechnology, more particularly, to water-soluble polymeric delivery systems for the imaging, evaluation and / or treatment of rheumatoid arthritis and other inflammatory diseases. Using modern MR imaging techniques, the specific accumulation of macromolecules in arthritic joints in adjuvant-induced arthritis in rats is demonstrated. The strong correlation between the uptake and retention of the MR contrast agent labeled polymer with histopathological features of inflammation and local tissue damage demonstrates the practical applications of the macromolecular delivery system of the invention.
Owner:BOARD OF RGT UNIV OF NEBRASKA
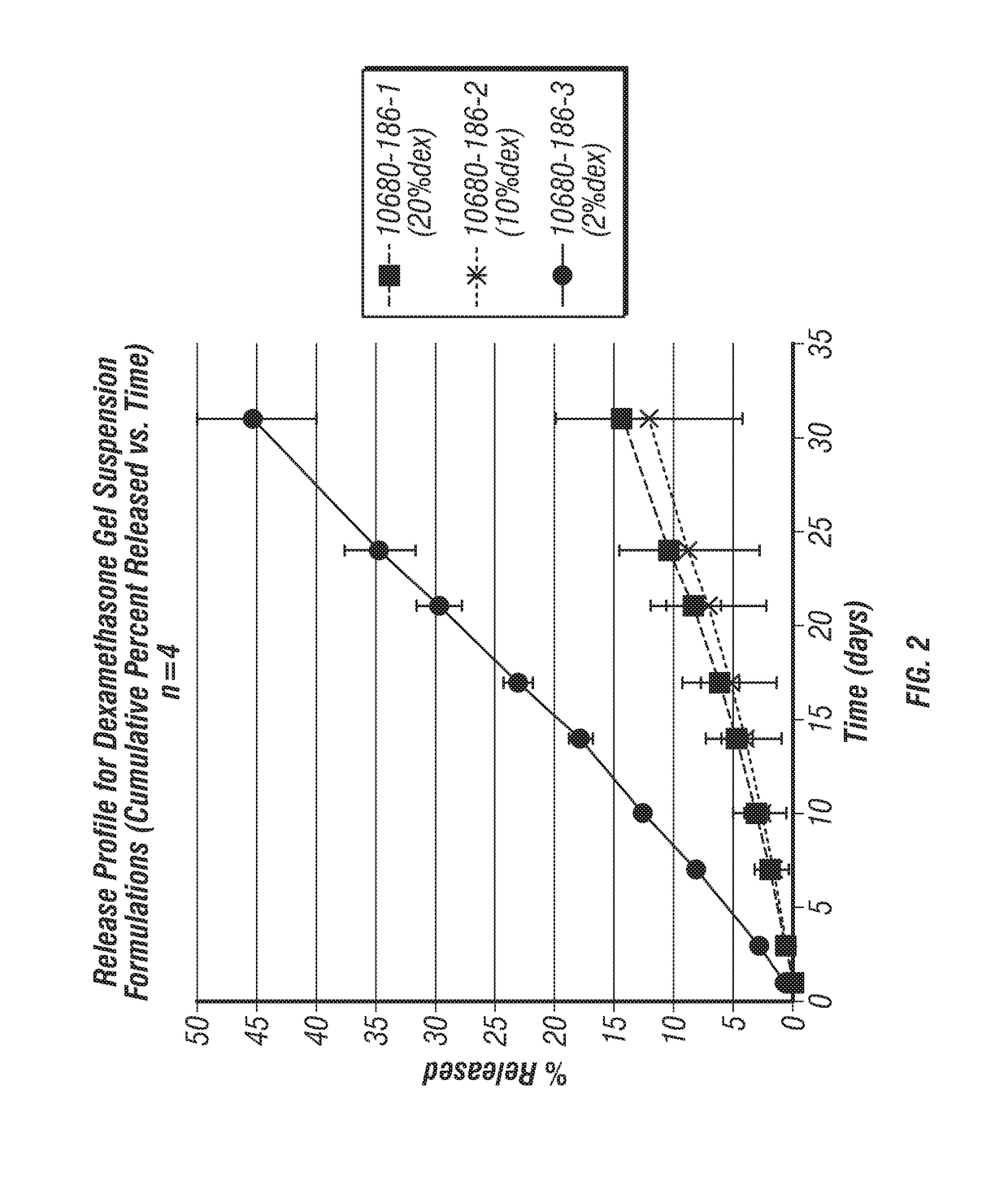
Extended release formulations for intra-articular applications
The present invention relates to a pharmaceutical composition comprising an aqueous suspension of: (i) crystalline N-(3,4-dichlorophenyl)-3-(pyridin-4-yl)-7-oxabicyclo[2.2.1]heptane- 2-carboxamide ora pharmaceutically acceptable salt thereof; and (ii) a surfactant comprising a water-soluble co-polymer characterized by > 5% solubility in water at 25 DEG C. The compositions provide extended releaseof N-(3,4-dichlorophenyl)-3-(pyridin-4-yl)-7-oxabicyclo[2.2.1] heptane- 2-carboxamide, and are suitable for intra-articular injection to a joint of a patient suffering from arthritis, joint injury orcartilage injury.
Owner:NOVARTIS AG
Cartilage regeneration using chondrocyte and tgf-beta
InactiveUS20190224248A1Peptide/protein ingredientsPharmaceutical delivery mechanismFactor iiFhit gene
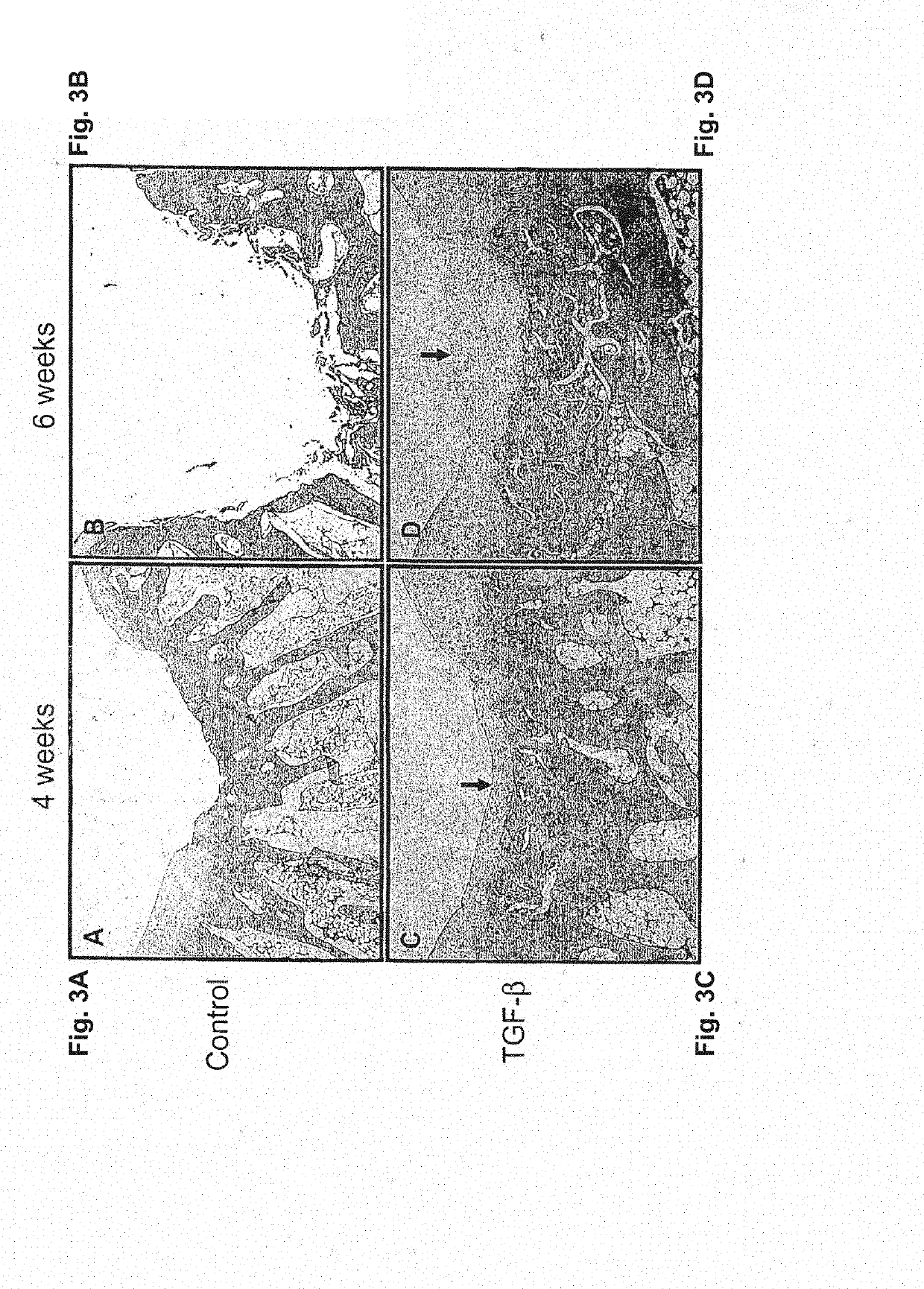
The present application is directed to a method of treating osteoarthritis, which includes obtaining a member of a transforming growth factor superfamily of proteins; obtaining a population of cultured mammalian cells that may contain vector encoding a gene, or a population of cultured connective tissue cells that do not contain any vector encoding a gene; and then transferring the protein and the connective tissue cells into an arthritic joint space of a mammalian host, such that the activity of the combination within the joint space results in regenerating connective tissue.
Owner:KOLON TISSUEGENE INC
Joint fat pad formulations, and methods of use thereof
ActiveUS10869955B2Good curative effectReduce exposureOrganic active ingredientsNervous disorderDiseaseJoint cavity
The present invention relates to formulations for administration to a joint fat pad of a subject, and to methods of treating joint pain, inflammation or disease. The disclosed formulations are intended for local administration to the joint fat pad to provide sustained release of a therapeutic agent to the joint cavity and surrounding tissues. The joint may be an arthritic joint, an injured joint or a surgically replaced joint. The therapeutic agent may be an analgesic agent, an anti-inflammatory agent or an immunosuppressive agent. A single administration of the formulation to the joint fat pad delivers a therapeutically effective amount of the therapeutic agent with reduced systemic exposure relative to a single systemic or a single intra-articular administration of a therapeutic dose of an identical therapeutic agent.
Owner:ALLERGAN INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com