CRISPR Cas9 system system for Bacillus subtilis genome edition and establishment method thereof
A Bacillus subtilis genome editing technology, applied in the field of genetic engineering, can solve problems such as bacterial infection and difficulty in controlling the fermentation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
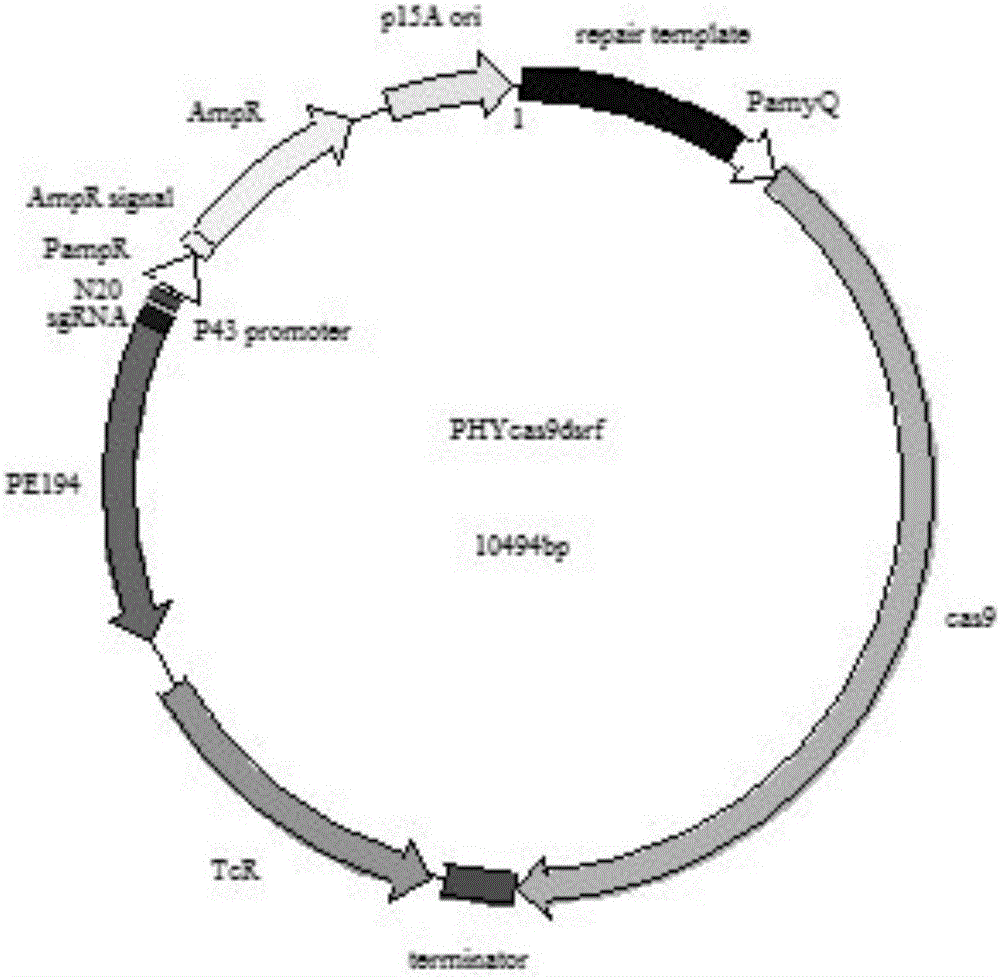
[0035] Example 1: Construction of CRISPR / Cas9 gene editing system to knock out plasmid PHY300dsrf
[0036] (1) According to the gene sequence of Bacillus subtilis, the sgRNA for specifically targeting the srfA-C gene is designed, and then the sgRNA double-stranded oligonucleotide sequence (such as SEQ ID NO: 2) is designed on the basis of the sgRNA of the srfA-C gene , look up the gene sequence of the PE194 temperature-sensitive replication origin (such as SEQ ID NO: 3) from NCBI and then synthesize sgRNA and PE194 sequences and connect them to the PMD18-T vector, using the following primers 5'-GGAACGTACAGACGCATTTTACATTTTTAGAAATGGGC-3' (such as SEQ ID NO: 5) Perform PCR amplification with 5'-CGTTTGTTGAACTACGCAGTCGGCTTAAACCAG-3' (such as SEQ ID NO: 6) to obtain Frag1. The reaction system is shown in Table 1.
[0037] Table 1 reaction system
[0038] 5xPhusion HF Reaction Buffer
[0039] The reaction procedure was as follows: pre-denaturation at 94°C for 4 min; 30 cy...
Embodiment 2
[0056] Embodiment 2: Bacillus subtilis transformation method
[0057] Dip the frozen Bacillus subtilis with an inoculation loop, then streak on the LB plate, and cultivate overnight at 37°C for activation. Pick a single colony and inoculate in 5mL LB liquid medium, and culture overnight at 37°C for 18h. Take a certain amount of overnight culture into 4.5mL of GMI, so that the OD600 value reaches 0.1-0.2, leaving 4.5mL of mixed bacterial liquid. Shake culture at 200 rpm at 37°C, measure OD600 every 20 minutes, when OD600 reaches 0.4-0.6 (about 60-90 minutes); continue shaking culture for 90 minutes, draw 0.05ml of bacterial liquid into a sterile test tube containing 0.45mL of preheated GMⅡ medium; shaking at 37°C for 90 minutes, at this time many competent cells formed in the culture; add 1 μg of plasmid (15-20uL), shake and culture at 37°C for 30 minutes; centrifuge to remove most of the supernatant, resuspend the cells, and spread on Incubate overnight at 37°C on a screenin...
Embodiment 3
[0062] Embodiment 3: use plasmid PHY300dsrf to knock out the srfA-C gene in B.subtilis168 genome
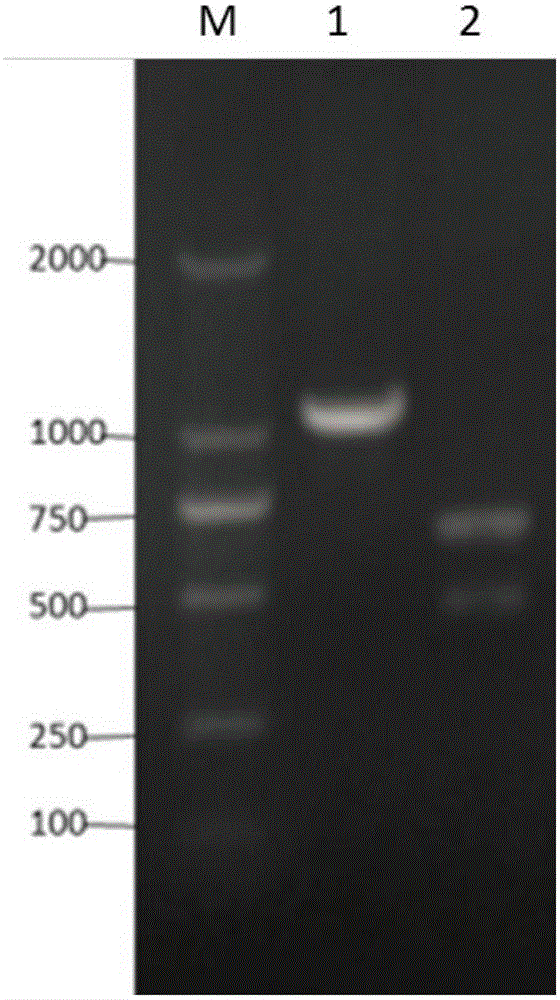
[0063]Using the method in Example 2, the knockout plasmid PHY300dsrf was transformed into competent cells of Bacillus subtilis, spread on LB solid medium (containing 20ug / mL tetracycline), and cultured overnight at 37°C. Pick positive clones, extract the genome, use the genome as a template, use the following primers 5'-CTCGAGGCTAGGGGCAGCGAGCAAACAGC-3' (such as SEQ ID NO: 17) and 5'-GAGCAGCTCTTTCGGCTCATAG-3' (such as SEQ ID NO: 18) to carry out PCR amplification, and then in Perform XbaI digestion verification at 37°C. When the srfA-C gene knockout strain performs genome homology repair, an XbaI restriction site is inserted at the srfA-C gene break, so it can be cut by the XbaI endonuclease, while the wild type will not be cut ( figure 2 , image 3 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com