SNP (Single Nucleotide Polymorphism) molecular marker related with back fat thickness trait of pig and detection method of SNP molecular marker
A technology of molecular markers and pig backfat thickness, applied in the field of molecular genetics, can solve the problems of lagging functional research and inaccurate gene positioning, etc., and achieve excellent economic value, low cost, and adapt to a wide range of pig breeds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
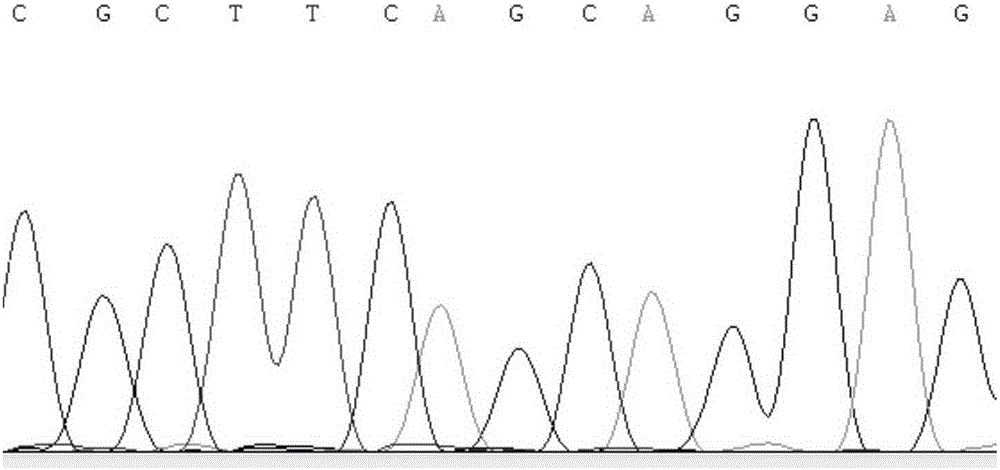
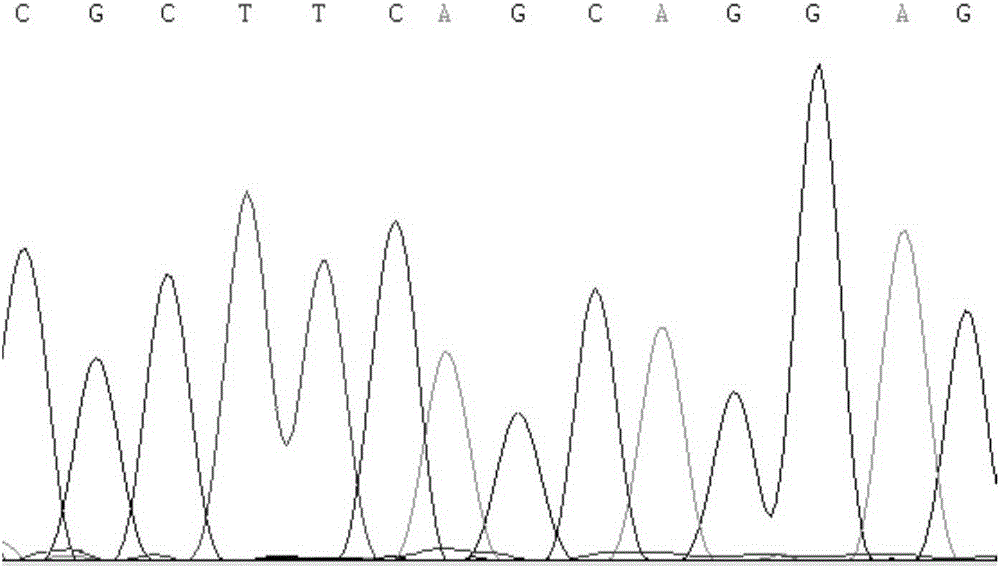
[0030] Example 1 Obtaining of SNP molecular markers related to pig backfat thickness traits
[0031] Taking 268 Duroc pigs with complete phenotype data as the research object, other Chinese local pig breeds (with no relatives among individuals) include 33 Jinhua pigs, 25 Neijiang pigs, 13 Erhualian pigs, 32 Wuzhishan pigs, 29 Bamei pigs and 26 Tibetan pigs are high-backfat breed groups; 30 Large White pigs and 29 Landrace pigs are low-backfat breed groups. For the above verification, some bases of the pig MARK4 gene segment were sequenced by PCR technology Sequences, supplemented with partial genome sequences.
[0032] According to the predicted porcine MARK4 gene sequence (XM_005658175) and human MARK4 gene sequence (NM_001199867) published by NCBI, primers were designed to amplify the porcine MARK4 gene mRNA sequence. fragments.
[0033] Table 1 MARK4 gene SNP detection specific primers
[0034]
[0035] According to reverse transcription PCR technology and RACE (Rapid-a...
Embodiment 2
[0037] Example 2 Establishment of a method for detecting SNP molecular markers related to pig backfat thickness traits
[0038] 1. Use RFLP restriction endonuclease to detect the population level of SNP sites
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com