CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-Cas9) homing sequences and primers thereof, and transgenic expression vector and establishment method thereof
A technology for guide sequences and expression vectors, applied in the fields of CRISPR-Cas9 guide sequence primers, transgene expression vectors and their construction, which can solve the problems of high cost, long preparation period, and many spliced fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
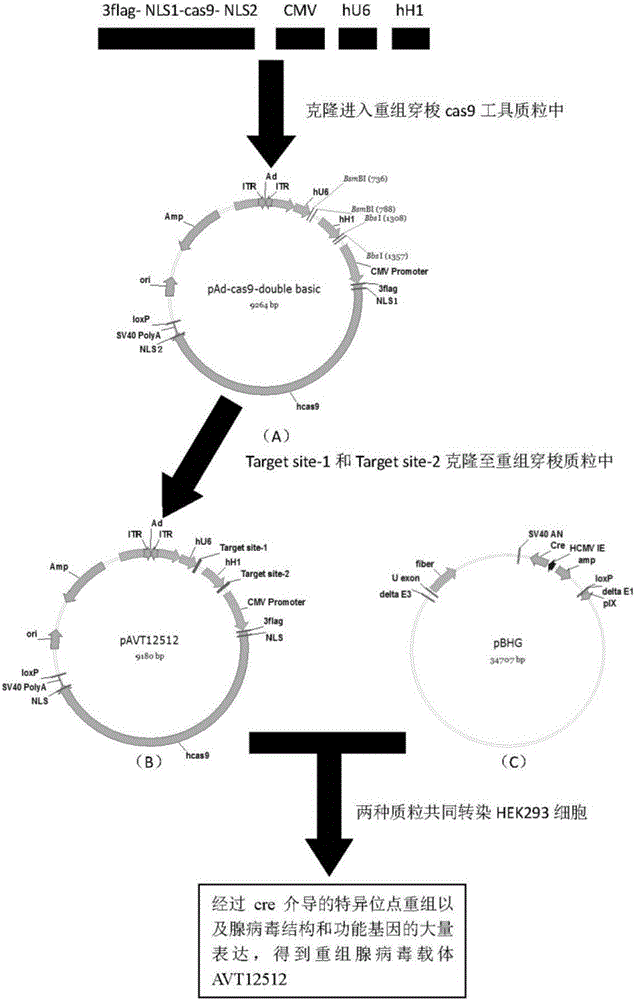
[0055] Example 1 Construction of recombinant adenovirus vector
[0056] 1. Materials
[0057] 1. Recombinant adenovirus backbone plasmid pBHG, adenovirus shuttle plasmid psb50, HEK293 cells, and homologous recombination enzyme are provided by Shiao (Shanghai) Biomedical Technology Co., Ltd.;
[0058] 2. Primers: According to the principles of primer design, the primers required for amplifying DNA fragments and target sites are designed. The primers are synthesized by Shanghai Jierui Bioengineering Co., Ltd., specifically:
[0059] Cas9-F: 5'-GTCAGATCCGCTAGCGCCACCATGGACTATAAGGACCACGACG-3' (SEQ ID NO.10)
[0060] Cas9-R: 5'-TTGCTCGAAGTCGACTCATTTCTTTTTTCTTAGCTTGACC-3' (SEQ ID NO.11)
[0061] CMV-F: 5'-GCTTGGATCCATTAGGCGGCCGCGTGGATAAC-3' (SEQ ID NO.12)
[0062] CMV-R: 5'-CATGGTGGCGCTAGCGGATCTGACGGTTCACTAAACCA-3' (SEQ ID NO.13)
[0063] hU6-F: 5'-AGCTCTAGACTCGAGAAGGTCGGGCAGGAAGAGG-3' (SEQ ID NO.14)
[0064] hU6-R: 5'-TTCAGCTCCCTATAACTATTAATAACTAATGCATGGCGGT-3' (SEQ ID NO.15) ...
Embodiment 2
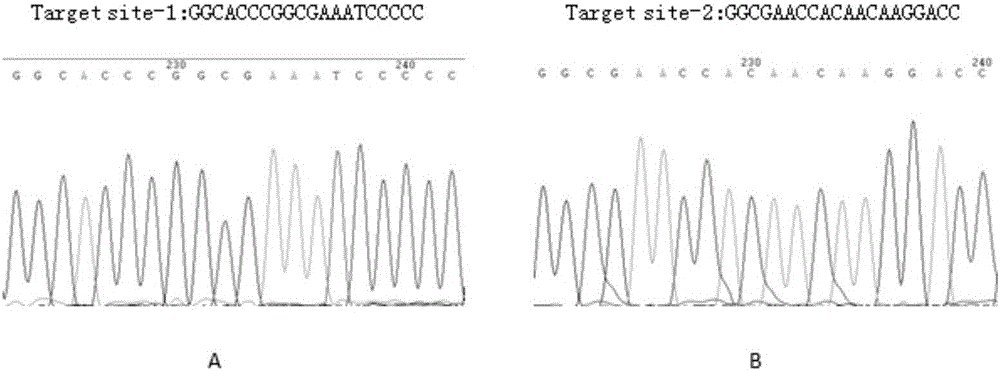
[0163] Example 2 Large-segment gene editing effect experiment of recombinant adenovirus vector AVT12512
[0164] 1: The object of action
[0165] HEK293T cells
[0166] Two: Experimental method
[0167] 1. The editing effect of the recombinant adenovirus vector AVT12512 of the present invention on large genome fragments of HEK293T cells.
[0168] (1) Take the HEK293T cell line out of liquid nitrogen, revive it in a 10cm dish, adjust the cell state to a normal growth state, and provide cells with a confluence of not more than 80%.
[0169] (2) One day before infection, HEK293T cells were digested with trypsin, the cells were resuspended and counted, and the cells were seeded in a 6-well plate and cultured at 5% CO2 at 37°C. Adjust the cells to a confluence of 40-50% before infection;
[0170] (3) Add the recombinant adenovirus vector AVT12512 according to MOI=80, mix the basic medium (serum-free) for the virus during infection, and add 6-8ug / ml polybrene at the same time, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com