MiR-205 gene knockout kit based on CRISPR-Cas9 gene knockout technology
A mir-205, mir-205-cas9 technology, applied in the field of genetic engineering, can solve the problem of ineffective inhibition of oncogenes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
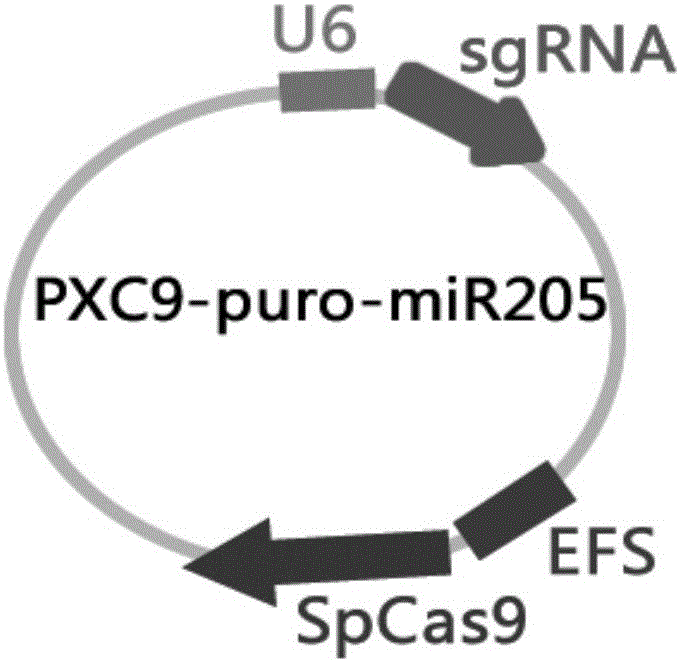
[0020] Embodiment 1: vector construction
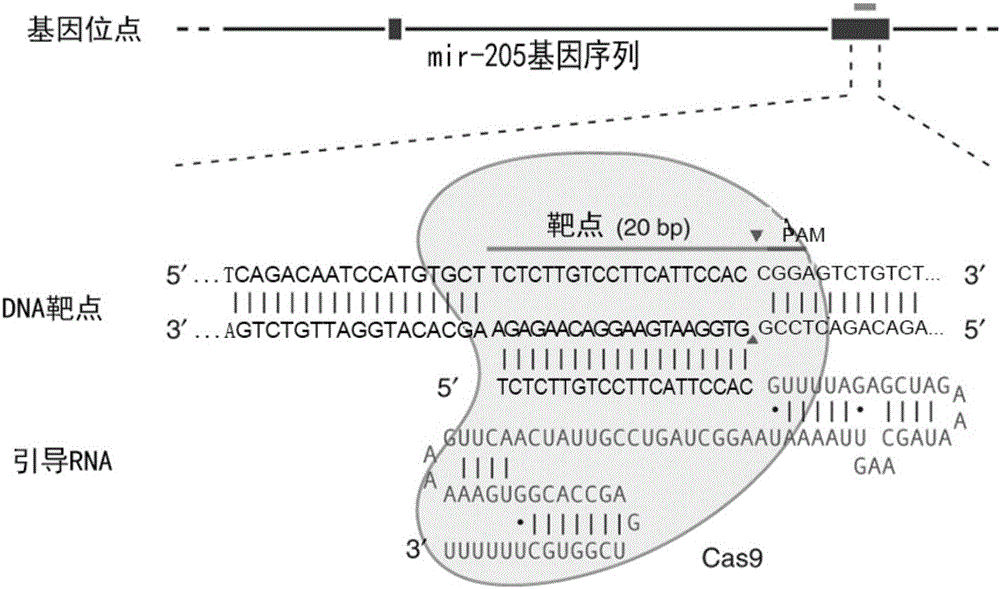
[0021] (1) Optimized design of miR-205 target
[0022] For the miR-205 gene (gene name MIR205, gene ID number: 406988, http: / / www.ncbi.nlm.nih.gov / gene / 406988), retrieve and download the miR-205 genome sequence (SEQ ID NO: 1):
[0023] 5'-AAAGATCCTCCAGACAATCCATGTGCTTCTCTTGTCCTTCATTCCACCGGAGTCTGTCTCATACCCAACCAGATTTCAGTGGAGTGAAGTTCAGGAGGCATGGAGCTGACA-3'
[0024]Use the online software FengZhanglab'sTargetFinder (http: / / crispr.mit.edu / ) and DNA2.0gRNA design tool software (https: / / www.dna20.com / eCommerce / cas9 / input) to design sgRNA, and input the above miR-205 Sequence, set and search to obtain the optimal target sequence. Select the target sequences recommended by both software, and select the optimal 4 target sequences, as shown in the following table:
[0025] Table 1
[0026] serial number target sequence Location SEQ ID NO:2 TCTCTTGTCCTTCATTCCAC 27-46 SEQ ID NO:3 CATACCCAACCAGATTTCAG 59-78 SEQ...
Embodiment 2
[0081] Example 2: Kit use and Knockout efficiency identification
[0082] (1) Plasmid transformation and amplification
[0083] 1) Take competent cells from a -80°C refrigerator and thaw in an ice box.
[0084] 2) Add 5uL of the ligation product to 50μL of DH5α competent cells in an ice bath, swirl gently to mix well, and bathe in ice for 30min.
[0085] 3) Heat shock in 42°C water bath for 90s (do not let the sample float).
[0086] 4) Quickly transfer the tube to an ice bath for 3 minutes.
[0087] 5) Add 500 μL SOC medium, mix well, put in a shaker, and activate at 200 rpm at 37° C. for 1 hour.
[0088] 6) Spread all on the LB plate containing Amp antibiotics, and place it at room temperature for 10 minutes until the liquid is absorbed. At the same time, add an appropriate amount of glass beads and shake back and forth to make the bacteria solution evenly coated. Invert the plate and transfer to a 37°C biochemical incubator for overnight culture.
[0089] 7) Pour 6 mL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com