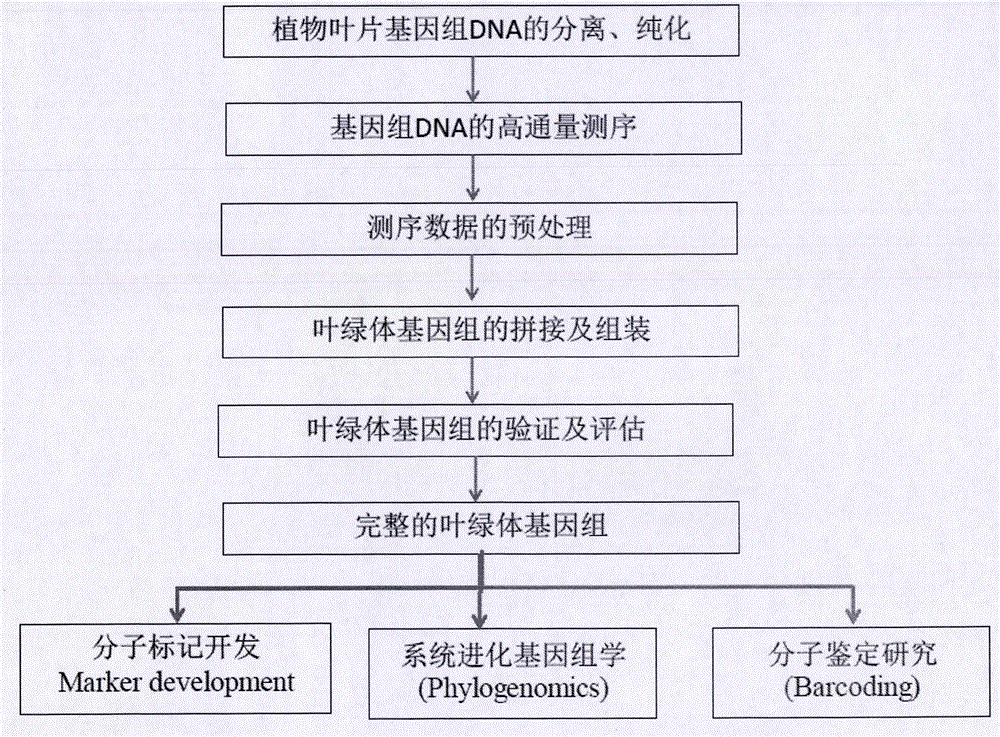
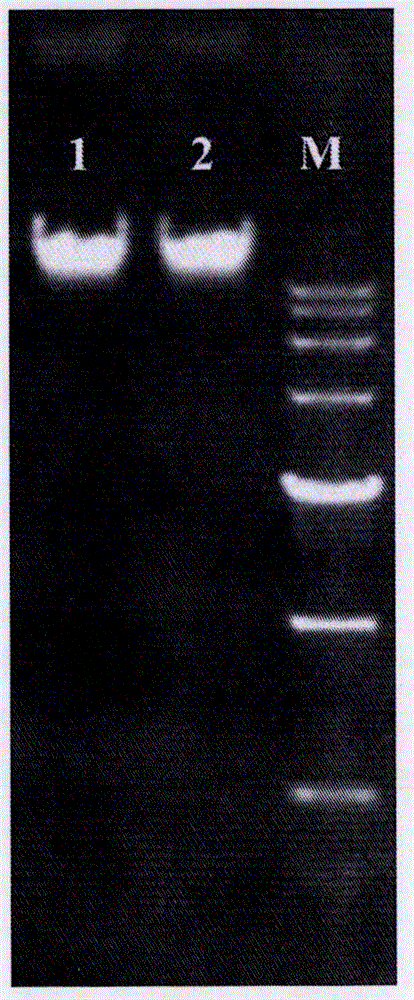
Simple, convenient, efficient and universal plant chloroplast genome sequencing method
A chloroplast genome and genome sequencing technology, applied in the field of biological sciences, can solve the problems of poor versatility, many operation steps, and long cycle, and achieve the effects of improving efficiency and versatility, reducing test steps, and improving assembly effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0040] Sequencing of the Chloroplast Genome of Eupatorium adenophorum
[0041] The genome size of Eupatorium adenophorum is about 1.6g. We extracted genomic DNA from the dark green leaves of Eupatorium adenophorum exposed to strong light for 2-3 days, and then used 5ug of the extracted DNA to construct a Solexa sequencing library, and then used the Hiseq2000 sequencing platform , PE100 for sequencing, and the sequencing data is 10G; then download all plant chloroplast genome sequences (ftp: / / ftp.ncbi.nlm.nih.gov / refseq / release / plastid) collected from NCBI as a reference sequence, and then high The data obtained by throughput sequencing is mapped using botwie software, and the reads that can be mapped are put into a new file, and finally the mapped reads data set is obtained. P1 and P2 total about 800Mb of data, and then use soapdenovo software The obtained reads were de novo assembled to obtain sequence contigs. A total of 3 scaffolds and 13 contigs were obtained, and the long...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com