Litopenaeus vannamei disease resistance related EST-SSR molecular marker and application thereof
A technology of molecular markers and disease-resistance-related genes, which is applied in the fields of climate change adaptation, DNA/RNA fragments, and microbial detection/inspection, can solve problems that have not been studied or reported, and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Discovery of SSR loci of functional genes related to disease resistance of Litopenaeus vannamei and design of primers for EST-SSR molecular markers
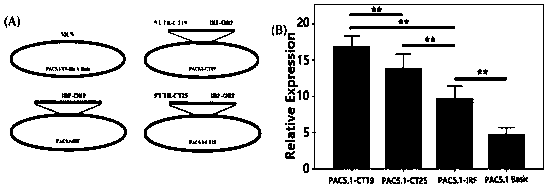
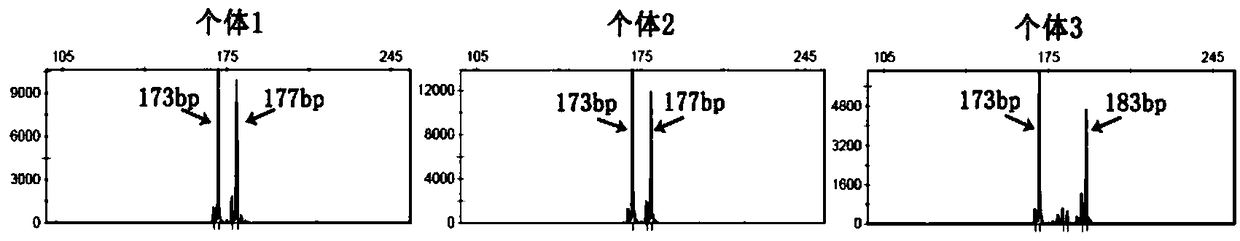
[0051] The inventor used the microsatellite search software SSR Hunter to analyze wxya Gene EST sequence to obtain core repeat sequence (CT) X , x = 10-30, when x = 19, the nucleotide sequence of the LvIRF-SSR is shown in SEQ ID NO.1.
[0052] Use the primer design software PrimerPrimer 5.0 to design primers for the upstream and downstream of the core repeat sequence of the above sequence, and obtain specific amplification containing the SSR site. wxya Specific amplification primers for gene 5'UTR sequence: LvIRF-5'UTR:
[0053] Plus strand 5'-ATCGGGATCCACTCGCAGAT-3' (SEQ ID NO: 2),
[0054] Negative strand 5'-GGCGACCTTAGACCGACGAG-3' (SEQ ID NO: 3); annealing temperature 56°C.
Embodiment 2
[0055] Example 2 Verification of the effect of SSR site polymorphisms on the expression of LvIRF at the cellular level
[0056] 1. Litopenaeus vannamei genomic DNA was extracted according to the instructions of OMEGA Animal Tissue DNA Extraction Kit. Use a Nanovue spectrophotometer to measure the DNA purity and concentration of the extracted DNA. The purity OD260 / 280 is 1.7~1.9, and the concentration is >100 ng / μl. Then it is detected by 1% agarose gel electrophoresis, and the integrity of the DNA fragment is detected by electrophoresis. Qualified DNA samples were stored at -20 °C until use.
[0057] 2. The total RNA of Litopenaeus vannamei was extracted according to the instructions of QIAGEN Animal Tissue RNA Extraction Kit. The purity and concentration of RNA were determined according to the above method, and the purity OD260 / 280 was 1.9-2.0. Qualified RNA samples were synthesized into cDNA using the Takara reverse transcription kit, and the operation was performed accord...
Embodiment 3
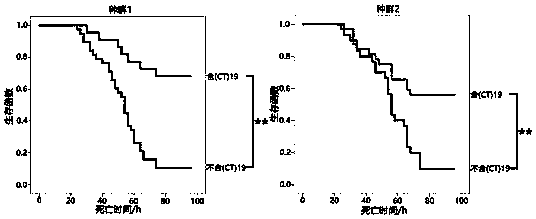
[0071] Example 3 Effect of LvIRF SSR site polymorphism on shrimp mortality
[0072] 1. Collect 120 prawns from two different sources from the breeding base, put them in the culture tank for 24 hours, and inject 10 μl of WSSV virus solution after the prawns are in a stable state (the content of WSSV virus is 10 5 copies / μl), the dead shrimps were collected every 4 h after the challenge, the dead shrimp within 96 h after the challenge were marked as group A, and the surviving and stable shrimp after 96 h after the challenge were marked as group B. The genomic DNA of prawns was extracted and the purity and concentration were detected according to the above method.
[0073] 2. Use the primer design software PrimerPrimer 5.0 to design primers for the upstream and downstream of the core repeat sequence of the above sequence, and obtain wxya SSR site primer nucleotide sequence: LvIRF-SSR:
[0074] Plus strand 5'-GATCCACTCGCAGATACAGAT-3' (SEQ ID NO: 8),
[0075] Negative strand...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com