Microsatellite fluorescence multi-PCR method for testing paternity of culter alburnus basilewsky
A technology for paternity identification and tuna, which is applied in the fields of biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effects of high experimental efficiency, saving DNA samples, and simple detection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] (1) Select the 3-year-old C. chinensis with mature gonads for artificial induction, and establish 12 full-sibling single-pair mating families; collect all parent individuals (24 in total) and some offspring individuals (20 for each family) Splashes, 480 in total);
[0027] (2) Take the fin ray of the Culturus chinensis individual, use the phenol-chloroform method to extract the genomic DNA, measure the concentration of the obtained DNA, and then dilute it to about 50 ng / μl;
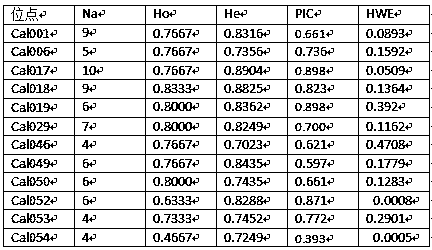
[0028] (3) Screening of polymorphic microsatellite sequences
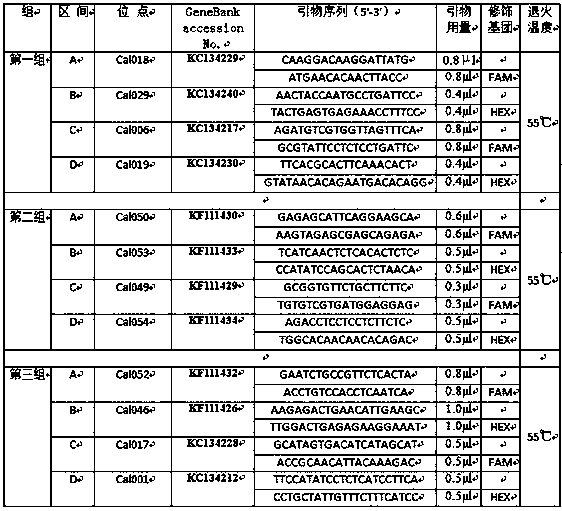
[0029] According to the NCBI sequence recorded in the NCBI, the Primer Premier 6.0 software was used to design the primer sequence of the Cichlid chinensis and the existing literature records, and to ensure that there is a large difference in the length of the target fragment (100-500bp), the primer sequence was sent to the biological company for synthesis. Using the genomic DNA obtained in step (1) as a template, the synthesized prime...
Embodiment 2
[0041] (1) Collect 32 cultured individuals in the Huzhou farm;
[0042] (2) Use the phenol-chloroform method to extract the DNA of each sample, and then dilute the obtained DNA to about 50 ng / μl after measuring its concentration;
[0043] (3) Synthesize primers according to the 12 kinds of primer sequences screened above and the modification method, the method is the same as that in Example 1;
[0044] (4) Perform multiple PCR amplification using the aforementioned three primer combinations, and the multiple PCR amplification system and amplification conditions are the same as in Example 1;
[0045] (5) After the PCR amplification product is properly diluted, it is mixed with a molecular weight internal standard and analyzed by capillary electrophoresis on an ABI 3130 sequencer;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com