Method for developing SSR (Simple Sequence Repeat) molecular mark of nibea albiflora for population identification
A technology of molecular marker and identification method, which is applied in the field of population molecular genetics of yellow croaker, achieves the effects of reduced cost, low experimental cost, and stable and reliable method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The following will clearly and completely describe the technical solutions in the embodiments of the present invention with reference to the accompanying drawings in the embodiments of the present invention. Obviously, the described embodiments are only some, not all, embodiments of the present invention. Based on the embodiments of the present invention, all other embodiments obtained by persons of ordinary skill in the art without making creative efforts belong to the protection scope of the present invention.
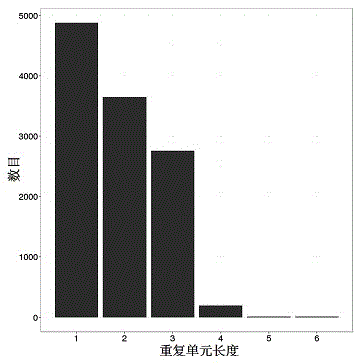
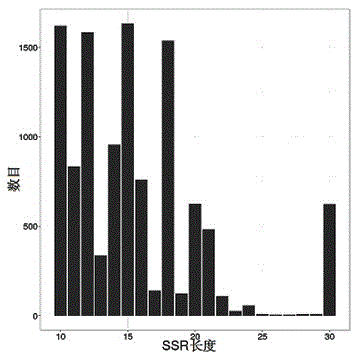
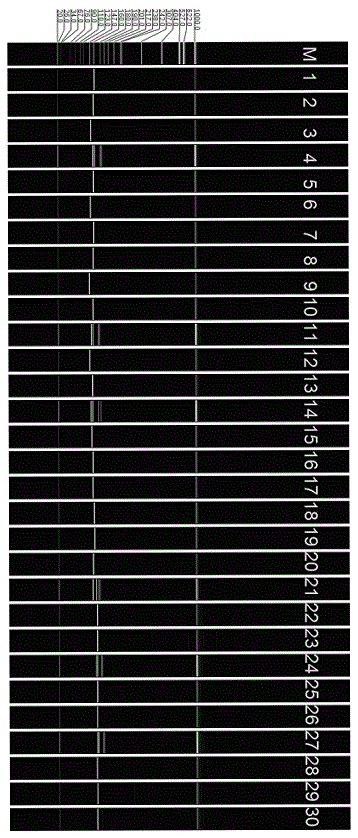
[0031] see Figure 1~4 , in an embodiment of the present invention, a method for developing croaker SSR molecular markers for population identification, specifically comprising the following steps:
[0032](1) Yellow croaker samples: The samples of croaker croaker came from the breeding base of croaker croaker in Ningde, Fujian. In order to obtain the gene sequences of different stages, this experiment used different developmental stages (fertilized eggs, newl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com