MUC13 gene determining susceptibility/resistance of piglet F4ac diarrhea and application thereof
A susceptibility and genetic technology, applied in the field of animal genetics and breeding, can solve the problems of no major breakthrough, no polymorphism, and inability to explain the genetic basis of ETCF4ac
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
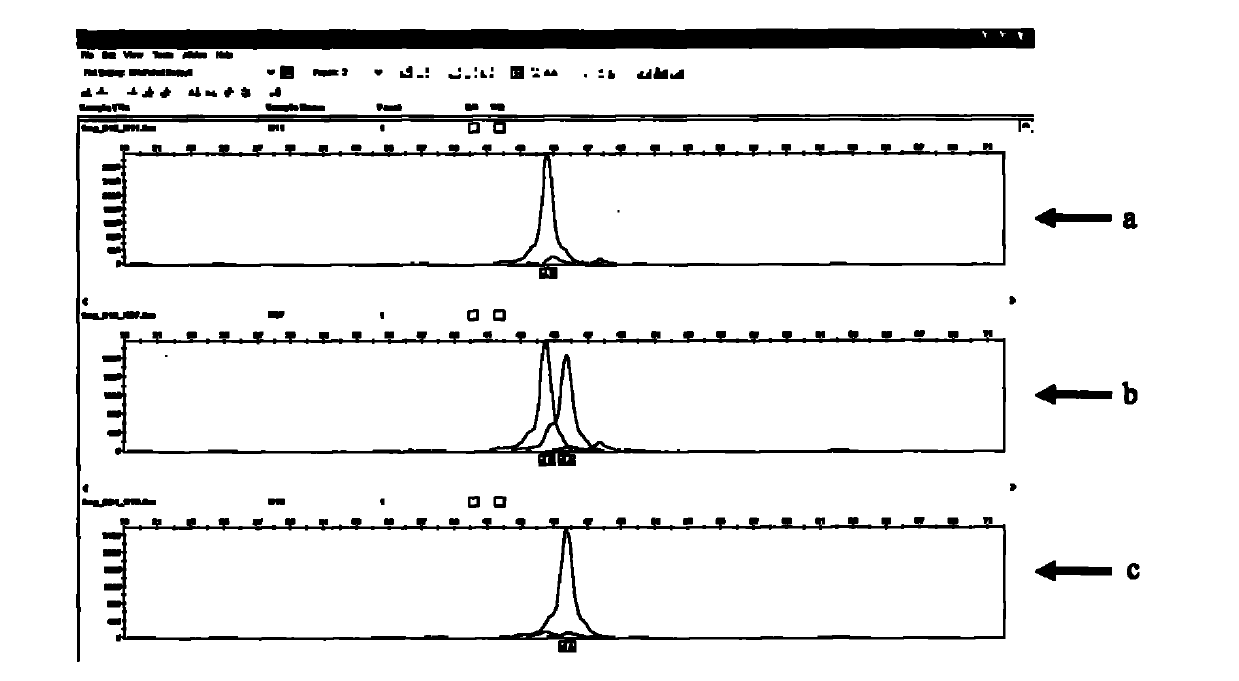
[0066] 144 purebred piglets including Duroc, Landrace and Large White pigs were used as experimental animals. These pigs were purchased from 5 large-scale commercial pig farms. Each pig breed selected at least 3 male lines, and each family selected 3-4 A 1-2 month old individual. After the 144 purebred piglets were slaughtered, the brush border ETEC F4ac adhesion phenotype test of porcine small intestinal epithelial cells was carried out in vitro, and the experimental methods and criteria were as described above.
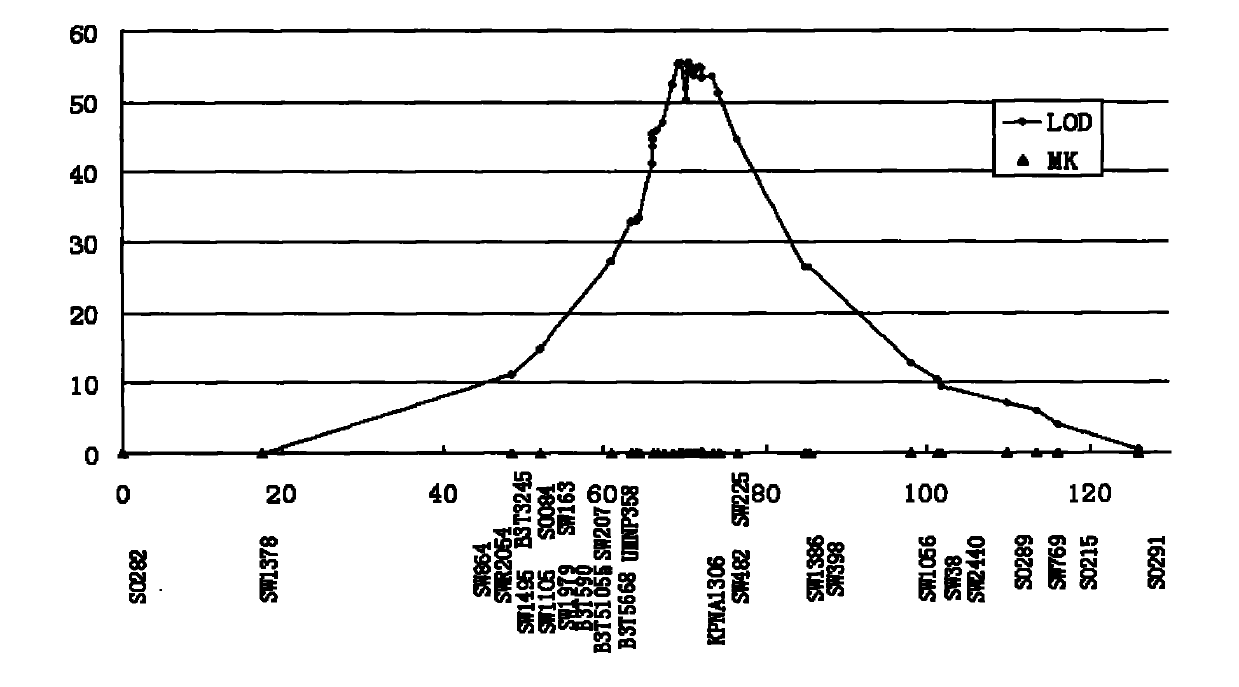
[0067] The SNPA37617G, SNPA32813G, SNPA42784C, SNPA46531G, and SNP C53019T loci of the MUC13 gene were subjected to SNP genotyping in the above 144 purebred piglets using the above-mentioned SNaPshot method. The amplification primers are shown in Table 4. The correlation statistical analysis was carried out between the genotype of each SNP site and the phenotype of ETEC F4ac adhered to pig intestinal brush border in vitro (see Table 5).
[0068] Table 4 is used to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com