Noninvasive Diagnosis of Fetal Aneuploidy by Sequencing
a fetal aneuploidy and sequencing technology, applied in the field of molecular diagnostics, can solve the problems of imposing small but potentially significant risks to both the fetus and the mother, limited reliability of non-invasive screening of fetal aneuploidy using maternal serum markers and ultrasound, and difficulty in measuring aneuploidy, so as to achieve more robust and statistically significant results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Subject Enrollment
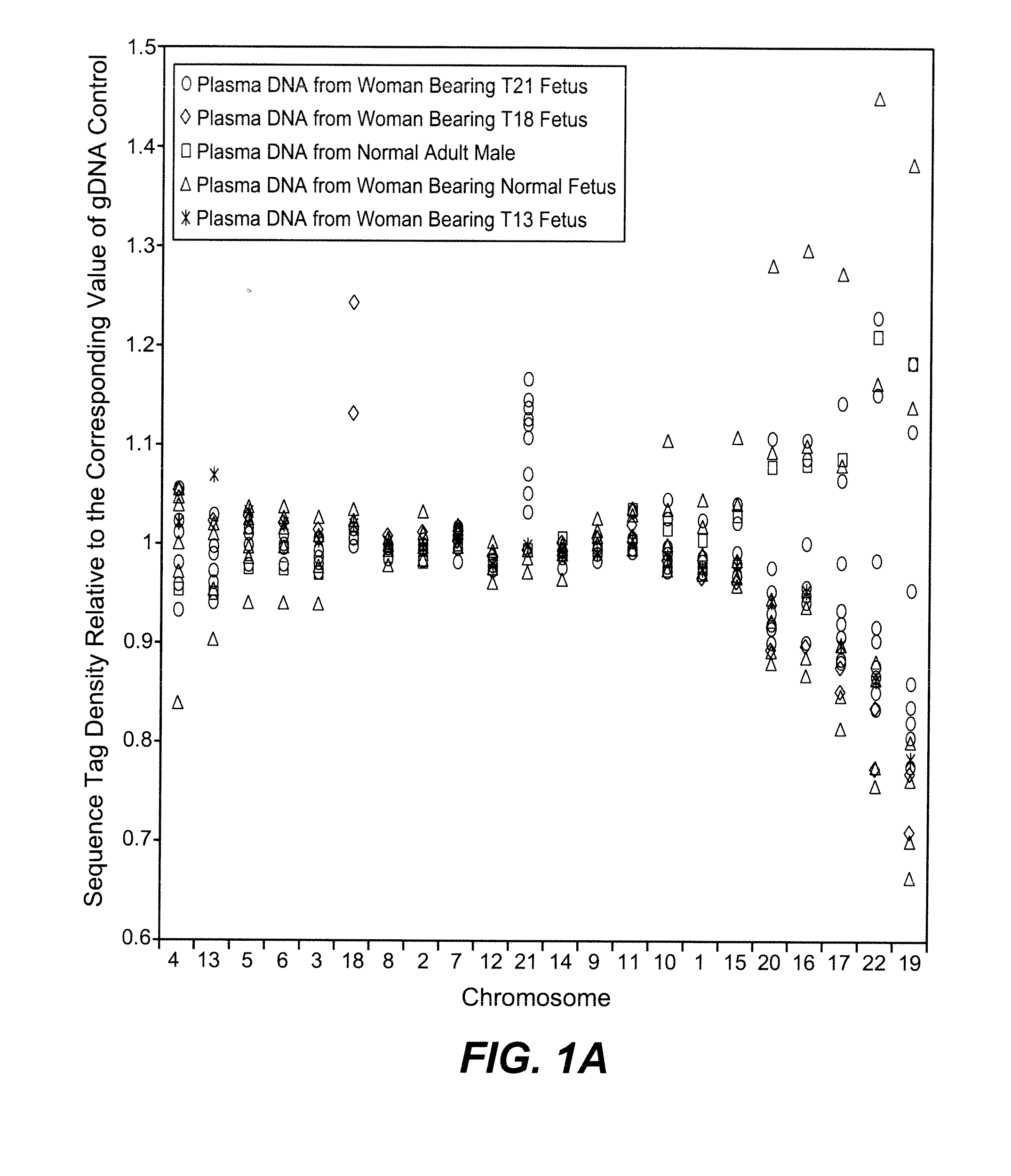
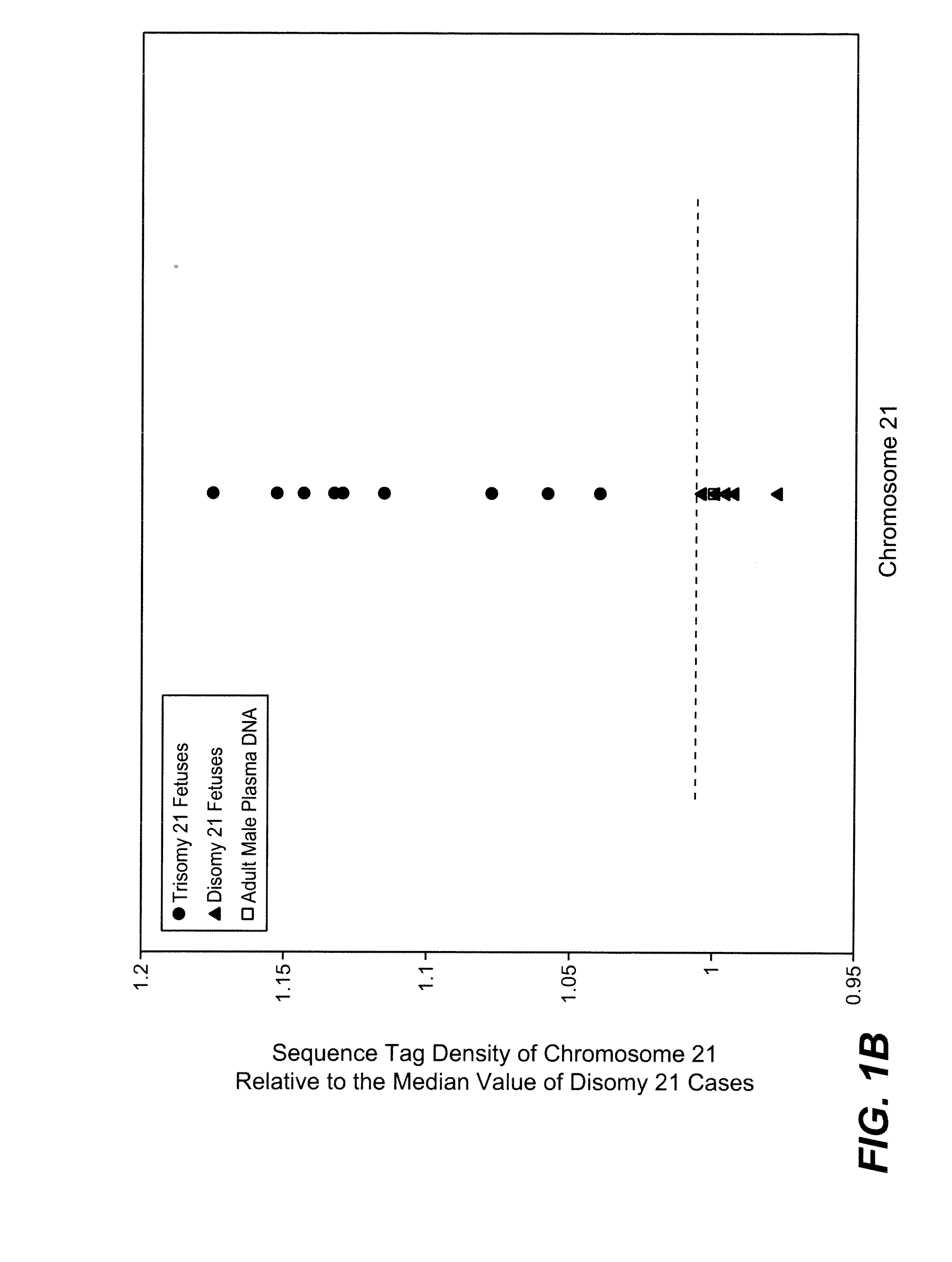
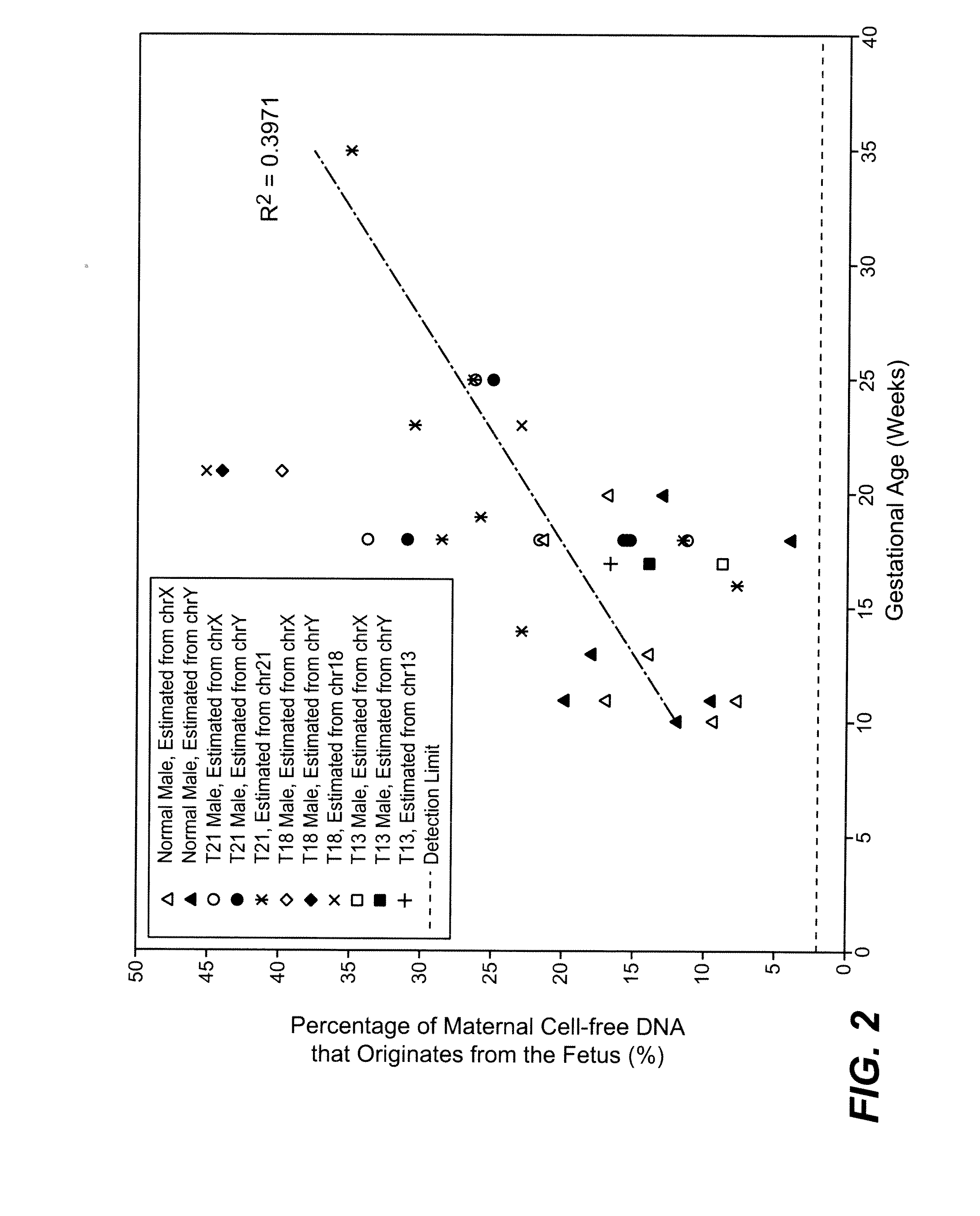
[0084]The study was approved by the Institutional Review Board of Stanford University. Pregnant women at risk for fetal aneuploidy were recruited at the Lucile Packard Children Hospital Perinatal Diagnostic Center of Stanford University during the period of April 2007 to May 2008. Informed consent was obtained from each participant prior to the blood draw. Blood was collected 15 to 30 minutes after amniocentesis or chorionic villus sampling except for 1 sample that was collected during the third trimester. Karyotype analysis was performed via amniocentesis or chorionic villus sampling to confirm fetal karyotype. 9 trisomy 21 (T21), 2 trisomy 18 (T18), 1 trisomy 13 (T13) and 6 normal singleton pregnancies were included in this study. The gestational age of the subjects at the time of blood draw ranged from 10 to 35 weeks (Table 1). Blood sample from a male donor was obtained from the Stanford Blood Center.
example 2
Sample Processing and DNA Quantification
[0085]7 to 15 ml of peripheral blood drawn from each subject and donor was collected in EDTA tubes. Blood was centrifuged at 1600 g for 10 minutes. Plasma was transferred to microcentrifuge tubes and centrifuged at 16000 g for 10 minutes to remove residual cells. The two centrifugation steps were performed within 24 hours after blood collection. Cell-free plasma was stored at −80 C until further processing and was frozen and thawed only once before DNA extraction. DNA was extracted from cell-free plasma using QIAamp DNA Micro Kit (Qiagen) or NucleoSpin Plasma Kit (Macherey-Nagel) according to manufacturers' instructions. Genomic DNA was extracted from 200 μl whole blood of the donors using QIAamp DNA Blood Mini Kit (Qiagen). Microfluidic digital PCR (Fluidigm) was used to quantify the amount of total and fetal DNA using Taqman assays targeting at the EIF2C1 locus on chromosome 1 (Forward: 5′ GTTCGGCTTTCACCAGTCT 3′ (SEQ ID NO: 1); Reverse: 5′ C...
example 3
Sequencing
[0086]A total of 19 cell-free plasma DNA samples, including 18 from pregnant women and 1 from a male blood donor, and genomic DNA sample from whole blood of the same male donor, were sequenced on the Solexa / Illumina platform. ˜1 to 8 ng of DNA fragments extracted from 1.3 to 5.6 ml cell-free plasma was used for sequencing library preparation (Table 1). Library preparation was carried out according to manufacturer's protocol with slight modifications. Because cell-free plasma DNA was fragmented in nature, no further fragmentation by nebulization or sonication was done on plasma DNA samples.
[0087]Genomic DNA from male donor's whole blood was sonicated (Misonix XL-2020) (24 cycles of 30 s sonication and 90 s pause), yielding fragments with size between 50 and 400 bp, with a peak at 150 bp. ˜2 ng of the sonicated genomic DNA was used for library preparation. Briefly, DNA samples were blunt ended and ligated to universal adaptors. The amount of adaptors used for ligation was 50...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Digital information | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com